5 releases
| 0.1.5 | Jan 19, 2025 |
|---|---|
| 0.1.4 | Nov 18, 2024 |
| 0.1.3 | May 13, 2024 |
| 0.1.2 | May 1, 2023 |
| 0.0.12 | Oct 29, 2022 |
#17 in Database implementations
553 downloads per month
6MB
5.5K
SLoC

GSearch: Ultra-fast and Scalable Genome Search based on Various MinHash-like Metrics and HNSW
Quick install on Linux
### pre-combiled binary
wget https://github.com/jean-pierreBoth/gsearch/releases/download/v0.1.5/gsearch_Linux_x86-64_v0.1.5.zip
unzip gsearch_Linux_x86-64_v0.1.5.zip
chmod a+x ./gsearch_Linux_x86-64_v0.1.5/*
cd gsearch_Linux_x86-64_v0.1.5
./gsearch -h
### Install via bioconda (hmmsearch_rs and hnswcore not available for now)
conda install -c conda-forge -c bioconda gsearch
GSearch stands for genomic search.
This package (currently in development) compute MinHash-like signatures of bacteria and archaea (or virus and fungi) genomes and stores the id of bacteria and MinHash-like signatures in a Hnsw structure for searching of new request genomes. A total of ~50,000 to ~60,000 lines of highly optimized Rust code were provided in this repo and several other crates/libraries develped for this repo, such as kmerutils, probminhash, hnswlib-rs and annembed, see below for details. Some of the libraries are very popular and have been used about ~40 thousand times, see here.
This package is developped by Jean-Pierre Both jpboth for the software part and Jianshu Zhao for the genomics part. We also created a mirror of this repo on GitLab and Gitee (You need to log in first to see the content), just in case Github service is not available in some region.
gsearch -h
************** initializing logger *****************
Approximate nearest neighbour search for microbial genomes based on MinHash-like metric
Usage: gsearch [OPTIONS] [COMMAND]
Commands:
tohnsw Build HNSW graph database from a collection of database genomes based on MinHash-like metric
add Add new genome files to a pre-built HNSW graph database
request Request nearest neighbors of query genomes against a pre-built HNSW graph database/index
ann Approximate Nearest Neighbor Embedding using UMAP-like algorithm
help Print this message or the help of the given subcommand(s)
Options:
--pio <pio> Parallel IO processing
--nbthreads <nbthreads> nb thread for sketching
-h, --help Print help
-V, --version Print version
Key functions
Sketching of genomes/tohnsw, to build hnsw graph database
The objective is to use the Jaccard index as an accurate proxy of mutation rate or Average Nucleitide Identity(ANI) or Average Amino Acide Identity (AAI) According to equation (Poisson model or Binomial model): $$ANI=1+\frac{1}{k}log\frac{2*J}{1+J}$$
or
$$ANI=(\frac{2*J}{1+J})^{\frac{1}{k}}$$
where J is Jaccard-like index (e.g. Jp from ProbMinHash or J from SuperMinHash, SetSketch or Densified MinHash, a class of locality sensitive hashing algorithms, suitable for nearest neighbor search, see below) and k is k-mer size. To achieve this we use sketching. We generate kmers along genome DNA or amino acid sequences and sketch the kmer distribution encountered (weighted or not) in a file, see kmerutils. Then final sketch is stored in a Hnsw database See hnsw or hnsw. Additionally, for high dimension dataset (e.g., d>32, we use d around 10,000 for prokaryotic genomes), HubNSW can be used via controlling the hierarchy of HNSW so that only 1 layer is built but the hubs in the dataset will be the "highway centers" that are useful for speeding up nivagation/search. See details in the HubNSW paper. We observed an 20% improvment for HNSW graph building, and an 20% to 30% improvement in memory requirement and up to 75% speed improvement for HNSW graph search with the same accuracy in HNSW.
The sketching and HNSW graph database building is done by the subcommand tohnsw.
The Jaccard index come in 2 flavours:
1. The probability Jaccard index that takes into account the Kmer multiplicity. It is defined by :
$$J_{P(A,B)}=\sum_{d\in D} \frac{1}{\sum_{d'\in D} \max (\frac{\omega_{A}(d')}{\omega_{A}(d)},\frac{\omega_{B}(d')}{\omega_{B}(d)})}$$
where $\omega_{A}(d)$ is the multiplicity of $d$ in A
(see Moulton-Jiang-arxiv).
In this case for J_p we use the probminhash algorithm as implemented in probminhash, or see original paper/implementation here.
2. The unweighted (simple) Jaccard index defined by :
$$Jaccard(A,B)=\frac{A \cap B}{A \cup B}$$
In this case for J we use the SuperMinHash, SetSketch (based on the locality sensitivity in section 3.3 or joint maximum likelihood estimation in section 3.2, joint estimation) or densified MinHash based on One Permutation Hashing with Optimal Densification method or One Permutation Hashing with Faster Densification method, also implemented in probminhash mentioned above.
The above mentioned choices for sketching can be specified in "gsearch tohnsw" subcommand via the --algo option (prob, super/super2, hll and optdens). We suggest using either ProbMinHash or Optimal Densification. For SuperMinhash, 2 implementations are available: super is for floating point type sketch while super2 is for integer type sketch. The later is much faster than the former. hll is for SetSketch, we name it hll because the SetSketch data structure can be seen as a similar implementation of HyperLogLog, but adding new algorithms for similarity estimation (e.g., Locality Sensitivity, LSH or Joint Maximum Likelihood Estimation, JMLE for Jaccard index) in additional to distinct element counting. Also becasuse we think HyperLogLog is a beautiful name, which describes the key steps of the implementation.
The estimated Jaccard-like index is used to build HNSW graph database, which is implemented in crate hnswlib-rs.
The sketching of reference genomes and HNSW graph database building can take some time (less than 0.5 hours for ~65,000 bacterial genomes of GTDB for parameters giving a correct quality of sketching, or 2 to 3 hours for entire NCBI/RefSeq prokaryotic genomes. ~318K). Result is stored in 2 structures:
- A Hnsw structure storing rank of data processed and corresponding sketches.
- A Dictionary associating each rank to a fasta id and fasta filename.
The Hnsw structure is dumped in hnswdump.hnsw.graph and hnswdump.hnsw.data The Dictionary is dumped in a json file seqdict.json
gsearch tohnsw -h
************** initializing logger *****************
Build HNSW graph database from a collection of database genomes based on MinHash-like metric
Usage: gsearch-new tohnsw [OPTIONS] --dir <hnsw_dir> --kmer <kmer_size> --sketch <sketch_size> --nbng <neighbours> --algo <sketch_algo>
Options:
-d, --dir <hnsw_dir> directory for storing database genomes
-k, --kmer <kmer_size> k-mer size to use
-s, --sketch <sketch_size> sketch size of minhash to use
-n, --nbng <neighbours> Maximum allowed number of neighbors (M) in HNSW
--ef <ef> ef_construct in HNSW
--scale_modify_f <scale_modify> scale modification factor in HNSW or HubNSW [default: 1.0]
--algo <sketch_algo> specifiy the algorithm to use for sketching: prob, super/super2, hll or optdens/revoptdens
--aa --aa Specificy amino acid processing, require no value
--block --block : sketching is done concatenating sequences
-h, --help Print help
The HubNSW idea mentioned above can be achieved via a small scale_modify_f=0.25.
Adding genomes to existing/pre-built database
For adding new genomes to existing database, the add subcommand is being used. It will automatically load sketching files, graph files and also paremeters used for building the graph and then use the same parameters to add new genomes to exisiting database genomes.
************** initializing logger *****************
Add new genome files to a pre-built HNSW graph database
Usage: gsearch add --hnsw <hnsw_dir> --new <newdata_dir>
Options:
-b, --hnsw <hnsw_dir> set the name of directory containing already constructed hnsw data
-n, --new <newdata_dir> set directory containing new data
-h, --help Print help
Request, search new genomes agains pre-built database
For requests the subcommand request is being used. It reloads the dumped files, hnsw and seqdict related takes a list of fasta files containing requests and for each fasta file dumps the asked number of nearest neighbours according to distance mentioned above.
************** initializing logger *****************
Request nearest neighbors of query genomes against a pre-built HNSW graph database/index
Usage: gsearch request --hnsw <DATADIR> --nbanswers <nb_answers> --query <request_dir>
Options:
-b, --hnsw <DATADIR> directory contains pre-built database files
-n, --nbanswers <nb_answers> Sets the number of neighbors for the query
-r, --query <request_dir> Sets the directory of request genomes
-h, --help Print help
A tabular file will be saved to disk in current directory (gsearch.neighbors.txt) with 3 key columns: query genome path, database genome path (ranked by distance) and distance. The distance can be transformed into ANI or AAI according to the equation above. We provide the program reformat (also parallel implementation) to do that:
reformat -h
Processes input files for ANI calculation
Usage: reformat <kmer> <model> <input_file> <output_file>
Arguments:
<kmer> The kmer value used for ANI calculation (16)
<model> The model to be used for ANI calculation (1 or 2,corresponding to Poisson model and Binomial model)
<input_file> File containing the data to be transformed into tabular format
<output_file> File where the tabular output will be saved
Options:
-h, --help Print help
-V, --version Print version
reformat 16 1 ./gsearch.neighbors.txt ./clean.txt
You will then see the clean output like this:
| Query_Name | Distance | Neighbor_Fasta_name | Neighbor_Seq_Len | ANI |
|---|---|---|---|---|
| test03.fasta.gz | 5.40E-01 | GCF_024448335.1_genomic.fna.gz | 4379993 | 97.1126 |
| test03.fasta.gz | 8.22E-01 | GCF_000219605.1_genomic.fna.gz | 4547930 | 92.5276 |
| test03.fasta.gz | 8.71E-01 | GCF_021432085.1_genomic.fna.gz | 4775870 | 90.7837 |
| test03.fasta.gz | 8.76E-01 | GCF_003935375.1_genomic.fna.gz | 4657537 | 90.5424 |
| test03.fasta.gz | 8.78E-01 | GCF_000341615.1_genomic.fna.gz | 4674664 | 90.4745 |
| test03.fasta.gz | 8.79E-01 | GCF_014764705.1_genomic.fna.gz | 4861582 | 90.4108 |
| test03.fasta.gz | 8.79E-01 | GCF_000935215.1_genomic.fna.gz | 4878963 | 90.398 |
| test03.fasta.gz | 8.82E-01 | GCF_002929225.1_genomic.fna.gz | 4898053 | 90.2678 |
| test03.fasta.gz | 8.83E-01 | GCA_007713455.1_genomic.fna.gz | 4711826 | 90.2361 |
| test03.fasta.gz | 8.86E-01 | GCF_003696315.1_genomic.fna.gz | 4321164 | 90.1098 |
Query_Name is your query genomes, Distance is genomic Jaccard distance (1-J/Jp), Neighbor_Fasta_name is the nearest neighbors based on the genomic Jaccard distance, ranked by distance. ANI is calculated from genomic Jaccard distance according the equaiton above between you query genome and nearest database genomes.
We also provide scripts for analyzing output from request and compare with other ANI based methods here: https://github.com/jianshu93/gsearch_analysis
SuperANI
Additional ANI calculation (if you do not want to use MinHash estimated ANI) for the query genomes and nearest neighbor genomes can be performed via the program superani:
superani -h
************** initializing logger *****************
Computing average nucleotide identity between reference and query genomes via sparse kmer chaining or Open Syncmer with Densified MinHash
Usage: superani --ql <FILE> --rl <FILE> --output <FILE>
Options:
-q, --ql <FILE> A file containing a list of query genome paths (.gz supported)
-r, --rl <FILE> A file containing a list of reference genome paths (.gz supported)
-o, --output <FILE> Output file to write results
-h, --help Print help
-V, --version Print version
The input is query genome path and reference genome path and output is ANI between each query and each reference genome. Both input files can be created by extracting from the output of gsearch request command with some modification of genome path. Multithreaded computation is supported and it can also be used as a seperate ANI calculator.
FragGeneScanRs
### predict genes for request at amino acid level, credit to original author but we rewrite the user inteface to be consistent with gsearch and other commands
FragGeneScanRs -s ./data/NC_000913.fna -o NC_000913 -t complete -p 8
hmmsearch_rs
hmmsearch_rs -h
************** initializing logger *****************
Search protein sequences against HMM profiles
Usage: hmmsearch_rs [OPTIONS] --faa <fasta> --hmm <hmm>
Options:
-f, --faa <fasta> Path to the faa file containing the protein sequences
-m, --hmm <hmm> Path to the HMM file
-o, --output <output> Output file to save the search results
-h, --help Print help
-V, --version Print version
we wrapped the HMMER C API and made modification so that the output is tabular for easy parsing. Universal gene HMMs can be found in the data folder. This command is only available in the release page for linux, not via bioconda. It is available for MacOS
### search a proteome agains a HMM pforile
hmmsearch_rs -f ./data/test03.faa -m ./data/HMM_bacteria/PF00380.20.HMM
SuperAAI
Calculate AAI between genomes based on FracMinhash
************** initializing logger *****************
Compute Average Amino Acid Identity (AAI) via FracMinHash/Sourmash for genomes
Usage: superaai [OPTIONS] --ql <FILE> --rl <FILE> --output <FILE>
Options:
-q, --ql <FILE> File containing list of query protein paths (.faa format, .gz supported)
-r, --rl <FILE> File containing list of reference protein paths (.faa format, .gz supported)
-o, --output <FILE> Output file to write results
-k, --kmer <INT> K-mer size for MinHash calculation [default: 7]
-l, --scaled <INT> Scaled factor for MinHash calculation [default: 100]
-s, --sketch <INT> Sketch size for MinHash (number of hashes) [default: 5120]
-h, --help Print help
-V, --version Print version
The input is query genome path (proteome) and reference genome path (proteome) and output is AAI between each query and each reference genome.
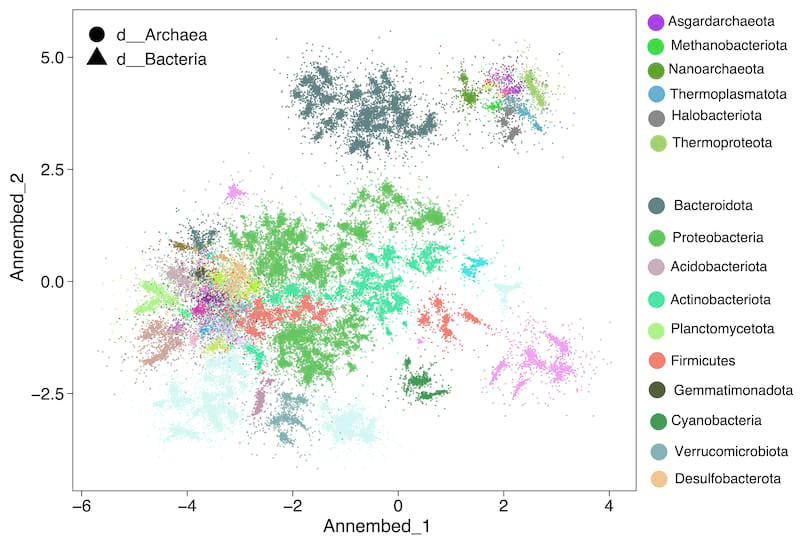
Ann
For UMAP-like algorithm to perform dimension reduction and then visuzlizing genome database, we run it after the tohnsw step (pre-built database) (see below useage ann section). See annembed crate for details. Then the output of this step can be visualized, for example for the GTDB v207 we have the following plot. See paper here.
Database split
We provide a bunch of scripts to allow split database genomes into N pieces and build HNSW graph database for each piece, then run search of new genomes against those pieces and collect results. This will only requires 1/N memory for your machine at the cost of additional sketching for the same query genomes.
### for a folder with genomes in it, we can split it into N subfolders randomly by running:
./scripts/split_folder.sh input_folder_path output_folder_path 3
### using the output from above split_folder.sh step, build database for each subfolder
./scripts/multiple_build.sh output_folder_path gsearch_db_folder
### using output from above multiple_build.sh step, search new genomes againt each database
./scripts/multiple_search.sh gsearch_db_folder_output new_genome_folder_path output
Database clustering via CoreSet construction for large databases
For real world datasets with billions of genomes, in additional to the random split idea mentioned above, approximate clustering via CoreSet can be used via the hnswcore command:
### for linux, you will also need to download the pre-compiled binary, for macOS brew install will be ok
wget https://github.com/jianshu93/coreset/releases/download/v0.1.0/hnswcore_Linux_x86-64_v0.1.0.zip
unzip hnswcore_Linux_x86-64_v0.1.0.zip
chmod a+x ./hnswcore
- First, we need to build a database via MinHash sketch and have nearest neighbors for each database genome.
gsearch --pio 4000 --nbthreads 60 tohnsw -d ../files -k 16 -s 18000 -n 128 --ef 1600 --algo optdens
- Second, we use the output from the step 1 to run Coreset clustering, --dir is the current directory in step 1.
hnswcore -h
************** initializing logger *****************
Approximate Clustering via Streaming CoreSet Construction
Usage: hnswcore --dir <dir> --fname <fname> --type <typename> [COMMAND]
Commands:
coreset CoreSet Construction
help Print this message or the help of the given subcommand(s)
Options:
-d, --dir <dir> directory contains HNSW database files
-f, --fname <fname> HNSW database file basename
-t, --type <typename> type for HNSW distance, e.f., f32, u32
-h, --help Print help
You can ask for a given number of cluters so that each cluster of genome sketches is smaller in size:
### if the optdens and revoptdens was used (see step 1), the output HNSW database can be clustered into 5 pieces like this
hnswcore --dir ./ --fname hnswdump --type f32 coreset --cluster 5
- Use the dictjsontocsv.ipynb python notebook in scripts folder to transfrom the cluster membership information id in HNSW to actual genome id.
Seqeunce search
We also provide general purpose sequence search via BigSig(BItsliced Genomic Signature Index), which can be used to quickly identify reads against a reference genome database.
************** initializing logger *****************
Large-scale Sequence Search with BItsliced Genomic Signature Index (BIGSIG)
Usage: bigsig [COMMAND]
Commands:
construct Construct a BIGSIG
query Query a BIGSIG on one or more fasta/fastq.gz files
identify Identify reads based on probability
help Print this message or the help of the given subcommand(s)
Options:
-h, --help Print help
-V, --version Print version
You can run the 2 steps to perform read identification like this:
### ref_file_example is path for reference genomes, see below
time bigsig construct -r ref_file_example.txt -b test -k 31 -mv 21 -s 10000000 -n 4 -t 24
time bigsig identify -b test.mxi -q ./test_data/SRR548019.fastq.gz -n output -t 24
input format:
### ref_file_example.txt
Salmonella_enterica_salamae_VIII_74-1880 ./refs/74_1880.fasta
Salmonella_enterica_salamae_b_Ar0560 ./refs/Ar0560.fasta
Salmonella_enterica_houtenae_Type ./refs/NCTC10401.fasta
Salmonella_bongori_Type-NCTC12419 ./refs/S_bongoriType_NCTC12419.fasta
Simple case for install
Pre-built binaries will be available on release page binaries for major platforms (no need to install but just download and make it executable). We recommend you use the linux one (gsearch_Linux_x86-64_v0.1.5.zip) for linux system in this release page for convenience (only system libraries are required). For macOS, we recommend the universal binary mac-binaries (gsearch_Darwin_universal_v0.1.5.tar.gz). Or GSearch_pc-windows-msvc_x86-64_v0.1.5.zip for Windows.
Or if you have conda installed on linux
### we suggest python 3.8, so if you do not have one, you can create a new one
conda create -n python38 python=3.8
conda activate python38
conda config --add channels bioconda
conda install -c conda-forge -c bioconda gsearch
gsearch -h
or if you are on MacOS and have homebrew installed
brew tap jianshu93/gsearch
brew update
brew install gsearch
gsearch -h
Otherwise it is possible to install/compile by yourself (see install section)
### get the binary for linux (make sure you have recent Linux installed with GCC, e.g., Ubuntu 18.0.4 or above)
wget https://github.com/jean-pierreBoth/gsearch/releases/download/v0.1.5/gsearch_Linux_x86-64_v0.1.5.zip --no-check-certificate
unzip gsearch_Linux_x86-64_v0.1.5.zip
cd gsearch_Linux_x86-64_v0.1.5
## get the binary for macOS (universal)
wget https://github.com/jean-pierreBoth/gsearch/releases/download/v0.1.5/gsearch_Darwin_universal_v0.1.5.tar.gz --no-check-certificate
tar -xzvf gsearch_Darwin_universal_v0.1.5.tar.gz
## get the binary for Windows, ann subcommand is not available for windows for now
wget https://github.com/jean-pierreBoth/gsearch/releases/download/v0.1.3-beta/GSearch_pc-windows-msvc_x86-64_v0.1.3.zip
## Note that for MacOS, you need sudo previlege to allow external binary being executed
* **make it excutable (changed it accordingly on macOS)**
chmod a+x ./gsearch
### put it under your system/usr bin directory (/usr/local/bin/ as an example) where it can be called
mv ./gsearch /usr/local/bin/
### check install
gsearch -h
### check install MacOS, you may need to change the system setup to allow external binary to run by type the following first and use your admin password
sudo spctl --master-disable
Usage
##We then give here an example of utilization with prebuilt databases.
### download neighbours for each genomes (fna, fasta, faa et.al. are supported) as using pre-built database, probminhash or SetSketch/hll database (prob and hll)
wget http://enve-omics.ce.gatech.edu/data/public_gsearch/GTDBv207_v2023.tar.gz
tar -xzvf ./GTDBv207_v2023.tar.gz
### Densified MinHash database (optdens) for NCBI/RefSeq, go to https://doi.org/10.6084/m9.figshare.24617760.v1
### Download the file in the link by clicking the red download to you machine (file GSearch_GTDB_optdens.tar.gz will be downloaded)
wget http://enve-omics.ce.gatech.edu/data/public_gsearch/GSearch_optdens.tar.gz
mkdir GTDB_optdens
mv GSearch_GTDB_optdens.tar.gz GTDB_optdens/
### get test data, we provide 2 genomes at nt, AA and universal gene level
wget https://github.com/jean-pierreBoth/gsearch/releases/download/v0.0.12/test_data.tar.gz --no-check-certificate
tar xzvf ./test_data.tar.gz
###clone gsearch repo for the scripts
git clone https://github.com/jean-pierreBoth/gsearch.git
### test nt genome database
cd ./GTDB/nucl
##default probminhash database
tar -xzvf k16_s12000_n128_ef1600.prob.tar.gz
# request neighbors for nt genomes (here -n is how many neighbors you want to return for each of your query genome, output will be gsearch.neighbors.txt in the current folder)
gsearch --pio 100 --nbthreads 24 request -b ./k16_s12000_n128_ef1600_canonical -r ../../test_data/query_dir_nt -n 50
## reformat output to have ANI
../../gsearch/scripts/reformat.sh 16 1 ./gsearch.neighbors.txt ./clean_ANI.txt
## SetSketch/hll database
tar -xzvf k16_s4096_n128_ef1600.hll.tar.gz
gsearch --pio 100 --nbthreads 24 request -b ./k16_s4096_n128_ef1600_canonical_hll -r ../../test_data/query_dir_nt -n 50
## reformat output to have ANI
../../gsearch/scripts/reformat.sh 16 1 ./gsearch.neighbors.txt ./clean_ANI.txt
## Densified MinHash, download first, see above
cd GTDB_optdens
tar -xzvf GSearch_GTDB_optdens.tar.gz
cd GTDB
#nt
tar -xzvf k16_s18000_n128_ef1600_optdens.tar.gz
gsearch --pio 100 --nbthreads 24 request -b ./k16_s18000_n128_ef1600_optdens -r ../../test_data/query_dir_nt -n 50
## reformat output to have ANI
../../gsearch/scripts/reformat.sh 16 1 ./gsearch.neighbors.txt ./clean_ANI.txt
### or request neighbors for aa genomes (predicted by Prodigal or FragGeneScanRs), probminhash and SetSketch/hll
cd ./GTDB/prot
tar xzvf k7_s12000_n128_ef1600.prob.tar.gz
gsearch --pio 100 --nbthreads 24 request -b ./k7_s12000_n128_ef1600_gsearch -r ../../test_data/query_dir_aa -n 50
## reformat output to have AAI
../../gsearch/scripts/reformat.sh 16 1 ./gsearch.neighbors.txt ./clean_AAI.txt
#aa densified MinHash
cd ./GTDB_optdens
cd GTDB
tar -xzvf k7_s12000_n128_ef1600_optdens.tar.gz
gsearch --pio 100 --nbthreads 24 request -b ./k7_s12000_n128_ef1600_optdens -r ../../test_data/query_dir_aa -n 50
## reformat output to have AAI
../../gsearch/scripts/reformat.sh 16 1 ./gsearch.neighbors.txt ./clean_ANI.txt
### or request neighbors for aa universal gene (extracted by hmmer according to hmm files from gtdb, we also provide one in release page)
cd ./GTDB/universal
tar xzvf k5_n128_s1800_ef1600_universal_prob.tar.gz
gsearch --pio 100 --nbthreads 24 request -b ./k5_n128_s1800_ef1600_universal_prob -r ../../test_data/query_dir_universal_aa -n 50
###We also provide pre-built database for all RefSeq_NCBI prokaryotic genomes, see below. You can download them if you want to test it. Following similar procedure as above to search those much larger database.
##graph database based on probminhash and setsketc/hll, both nt and aa
wget http://enve-omics.ce.gatech.edu/data/public_gsearch/NCBI_RefSeq_v2023.tar.gz
##nt graph database based on densified MinHash, go to below link to download GSearch_k16_s18000_n128_ef1600_optdens.tar.gz
https://doi.org/10.6084/m9.figshare.24615792.v1
#aa graph database, densified MinHash, go to below link to download GSearch_k7_s12000_n128_ef1600_optdens.tar.gz
https://doi.org/10.6084/m9.figshare.22681939.v1
### Building database. database is huge in size, users are welcome to download gtdb database here: (<https://data.ace.uq.edu.au/public/gtdb/data/releases/release207/207.0/genomic_files_reps/gtdb_genomes_reps_r207.tar.gz>) and here (<https://data.ace.uq.edu.au/public/gtdb/data/releases/release207/207.0/genomic_files_reps/gtdb_proteins_aa_reps_r207.tar.gz>) or go to NCBI/RefSeq to download all available prokaryotic genomes
### build database given genome file directory, fna.gz was expected. L for nt and .faa or .faa.gz for --aa. Limit for k is 32 (15 not work due to compression), for s is 65535 (u16) and for n is 255 (u8)
gsearch --pio 2000 --nbthreads 24 tohnsw -d db_dir_nt -s 12000 -k 16 --ef 1600 -n 128 --algo prob
gsearch --pio 2000 --nbthreads 24 tohnsw -d db_dir_aa -s 12000 -k 7 --ef 1600 -n 128 --aa --algo prob
### When there are new genomes after comparing with the current database (GTDB v207, e.g. ANI < 95% with any genome after searcing, corresponding to >0.875 ProbMinHash distance), those genomes can be added to the database
### old .graph,.data and all .json files will be updated to the new one. Then the new one can be used for requesting as an updated database
gsearch --pio 100 --nbthreads 24 add -b ./k16_s12000_n128_ef1600_canonical -n db_dir_nt (new genomes directory)
### or add at the amino acid level, in the parameters.json file you can check whether it is DNA or AA data via the "data_t" field
cd ./GTDB/prot
gsearch --pio 100 --nbthreads 24 add -b ./k7_s12000_n128_ef1600_gsearch -n db_dir_nt (new genomes directory in AA format predicted by prodigal/FragGeneScanRs)
### visuzlizing from the tohnsw step at amino acid level (AAI distance), output order of genome files are the same with with seqdict.json
cd ./GTDB/prot
gsearch ann -b ./k7_s12000_n128_ef1600_gsearch --stats --embed
Output explanation
gsearch.neighbors.txt is the default output file in your current directory.
For each of your genome in the query_dir, there will be requested N nearest genomes found and sorted by distance (smallest to largest) or ANI (largest to smallest). If one genome in the query does not exist in the output file, meaning at this level (nt or aa), there is no such nearest genomes in the database (or distant away from the best hit in the database), you may then go to amino acid level or universal gene level.
Dependencies, features and Installation
Features
-
hnsw_rs relies on the crate simdeez to accelerate distance computation. On intel you can build hnsw_rs with the feature simdeez_f
-
annembed relies on openblas so you must choose between the features "annembed_openblas-static" , "annembed_openblas-system" or "annembed_intel-mkl". You may need to install gcc, gfortran and make. This can be done using the --features option as explained below, or by modifying the features section in Cargo.toml. In that case just fill in the default you want.
-
kmerutils provides a feature "withzmq". This feature can be used to store compressed qualities on a server and run requests. It is not necessary in this crate.
Install/compiling by yourself
First install Rust tools
curl --proto '=https' --tlsv1.2 -sSf https://sh.rustup.rs | sh
gsearch installation and compilation from Crates.io (not recommended)
- simple install without annembed feature
cargo install gsearch --no-default-features --features="simdeez_f"
- simple installation, with annembed enabled would be with intel-mkl
cargo install gsearch --no-default-features --features="annembed_intel-mkl,simdeez_f"
or with a system installed openblas:
cargo install gsearch --features="annembed_openblas-system,simdeez_f"
- On MacOS, which requires dynamic library link (you have to install openblas first):
(note that openblas install lib path is different on M1 MACs).
So you need to run:
brew install openblas
echo 'export LDFLAGS="-L/usr/local/opt/openblas/lib"' >> ~/.bash_profile
echo 'export CPPFLAGS="-I/usr/local/opt/openblas/include"' >> ~/.bash_profile
echo 'export PKG_CONFIG_PATH="/usr/local/opt/openblas/lib/pkgconfig"' >> ~/.bash_profile
source ~/.bash_profile
cargo install gsearch --features="annembed_openblas-system"
gsearch installation from the most recent version from github
- direct installation from github:
cargo install gsearch --features="annembed_intel-mkl" --git https://github.com/jean-pierreBoth/gsearch
- download and compilation
git clone https://github.com/jean-pierreBoth/gsearch
cd gsearch
#### build
cargo build --release --features="annembed_openblas-static"
cargo build --release --features="annembed_intel-mkl"
###on MacOS, which requires dynamic library link (install openblas first, see above):
cargo build --release --features="annembed_openblas-system"
Documentation generation
Html documentation can be generated by running (example for someone using the "annembed_openblas-system" feature):
cargo doc --features="annembed_openblas-system" --no-deps --open
Then install FragGeneScanRs
cargo install --git https://gitlab.com/Jianshu_Zhao/fraggenescanrs
Some hints in case of problem (including installing/compiling on ARM64 CPUs) are given here
Pre-built databases
We provide pre-built genome/proteome database graph file for bacteria/archaea, virus and fungi. Proteome database are based on genes for each genome, predicted by FragGeneScanRs (https://gitlab.com/Jianshu_Zhao/fraggenescanrs) for bacteria/archaea/virus and GeneMark-ES version 2 (http://exon.gatech.edu/GeneMark/license_download.cgi) for fungi.
- Bacteria/archaea genomes are the newest version of GTDB database (v207, 67,503genomes) (https://gtdb.ecogenomic.org), which defines a bacterial speces at 95% ANI. Note that GSearch can also run for even higher resolution species database such as 99% ANI.
- Bacteria/archaea genomes from NCBI/RefSeq until Jan. 2023 (~318,000 genomes)(https://www.ncbi.nlm.nih.gov/refseq/about/prokaryotes/), no clustering at a given ANI threshold.
- Virus data base are based on the JGI IMG/VR database newest version (https://genome.jgi.doe.gov/portal/IMG_VR/IMG_VR.home.html), which also define a virus OTU (vOTU) at 95% ANI.
- Fungi database are based on the entire RefSeq fungal genomes (retrived via the MycoCosm website), we dereplicated and define a fungal speices at 99.5% ANI.
- All four pre-built databases are available here:http://enve-omics.ce.gatech.edu/data/gsearch or at FigShare, see the download links here
Future developments considered are given here
References
-
Jianshu Zhao, Jean Pierre Both, Luis M Rodriguez-R, Konstantinos T Konstantinidis, GSearch: ultra-fast and scalable genome search by combining K-mer hashing with hierarchical navigable small world graphs, Nucleic Acids Research, 2024;, gkae609, https://doi.org/10.1093/nar/gkae609.
-
Jianshu Zhao, Jean Pierre Both, Konstantinos T Konstantinidis, Approximate nearest neighbor graph provides fast and efficient embedding with applications for large-scale biological data, NAR Genomics and Bioinformatics, Volume 6, Issue 4, December 2024, lqae172, https://doi.org/10.1093/nargab/lqae172
Dependencies
~44–84MB
~1.5M SLoC