9 releases
| 0.3.0 | Apr 20, 2025 |
|---|---|
| 0.2.6 | Apr 19, 2025 |
| 0.2.4 | Apr 24, 2024 |
| 0.2.3 | Apr 9, 2021 |
| 0.1.0 | Mar 3, 2021 |
#30 in Science
97 downloads per month
115KB
2K
SLoC
neuroformats
Handling of structural neuroimaging file formats for Rust.
The neuroformats crate provides access to structural neuroimaging data in Rust by implementing parsers for various file formats. The focus is on surface-based brain morphometry data, as produced from 3D or 4D magnetic resonance imaging (MRI) data by neuroimaging software suites like FreeSurfer, CAT12 and others.
Background
In surface-based neuroimaging, basically meshes representing the 3D structure of the human cortex are reconstructed from a segmented 3D brain image. Then properties of the human brain, like the thickness of the cortex at a specific position, are computed from the reconstruction and stored as per-vertex data for the mesh.
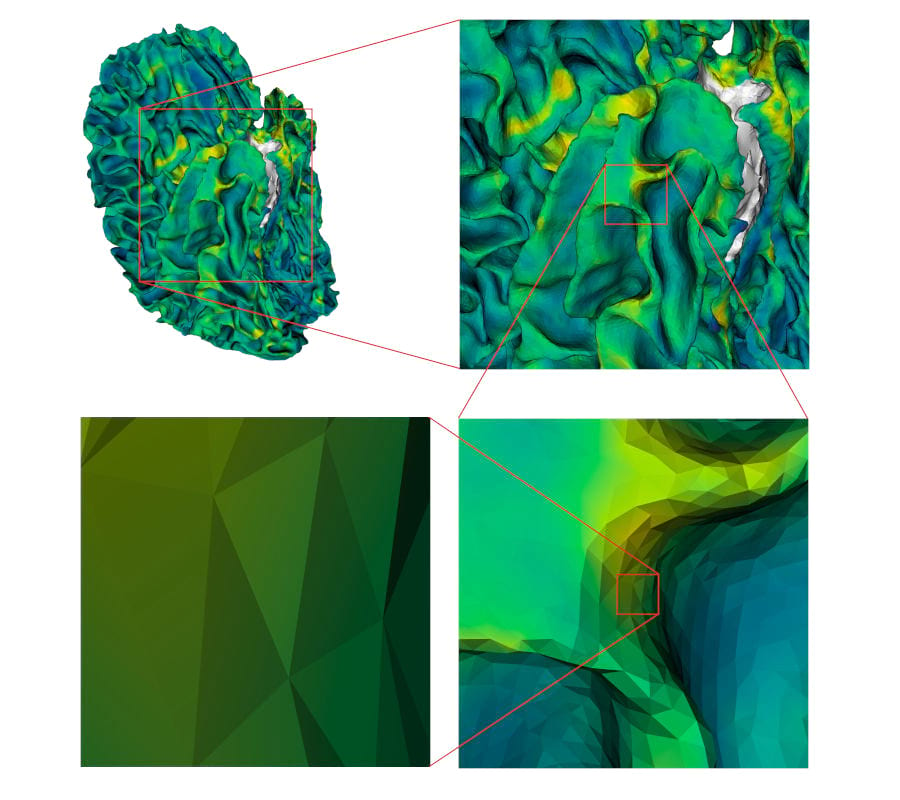
Fig. 1 A mesh representing the white surface of a human brain, with cortical thickness values mapped onto it using the viridis colormap. Different magnifications are displayed, and the triangles and vertex positions can be identified in the last one.
Installation
The neuroformats crate is on crates.io.
To use the library in your project, add it as a dependency in your Cargo.toml file, e.g.:
[dependencies]
neuroformats = "0.2.3"
Features
- Read and write FreeSurfer per-vertex data in curv format (like
subject/surf/lh.thickness): functionsneuroformats::read_curvandwrite_curv - Read and write brain meshes in FreeSurfer binary mesh format (like
subject/surf/lh.white):read_surfandwrite_surf - Read and write FreeSurfer label files (like
subject/label/lh.cortex.label):read_labelandwrite_label - Read FreeSurfer brain surface parcellations (a.k.a. brain atlas, like
subject/label/lh.aparc.annot):read_annot - Read and write FreeSurfer brain volumes and other data from MGH and MGZ files:
read_mghandwrite_mgh
Various utility functions are implemented for performing common computations on the returned structs, e.g. computing the vox2ras matrix from the MGH header data or finding all vertices in a brain surface parcellation that belong to a certain brain atlas region. The library can also export brain meshes in standard mesh formats (Wavefront Object Format, PLY format, glTF), optionally with vertex colors based on per-vertex data (from curv or MGH/MGZ files) and the viridis colormap for quick visualization in tools like MeshLab or Blender.
Documentation
API docs
The neuroformats API docs can be found at docs.rs/neuroformats.
Short usage example
Read vertex-wise cortical thickness computed by FreeSurfer:
use neuroformats::read_curv;
let curv = read_curv("subjects_dir/subject1/surf/lh.thickness").unwrap();
let thickness_at_vertex_0 : f32 = curv.data[0];
You now have a Vec<f32> with the cortical thickness values in curv.data. The order of the values matches the vertex order of the respective brain surface reconstruction (e.g., the white surface mesh of the left brain hemisphere in subjects_dir/subject1/surf/lh.white).
Full demo applications
- A simple example app that loads a brain mesh and per-vertex data (sulcal depth at each vertex), maps the per-vertex values to colors and exports the vertex-colored mesh in glTF format can be found in the ./examples/brain_export directory
- There is a small command line demo application that loads a brain surface mesh and raytraces an image based on the mesh to a PNG file available in the ./examples/brain_rpt directory. The demo uses the rpt crate by Eric Zhang and Alexander Morozov to do the raytracing. Instructions for building/running the demo application are at the top of the main.rs file.
- A simple demo that loads a brain mesh into bevy can be found in the ./examples/brain_bevy directory. This one requires some non-rust system dependencies to run, see the instructions in the main file for details.
See the neuroformats API docs and the unit tests in the source code for more examples.
Development Info
Please see the README_DEV file.
LICENSE
The neuroformats crate is free software, dual-licensed under the MIT or APACHE-2 licenses.
Contributions
Contributions are very welcome. Please get in touch before making major changes to avoid wasted effort.
Help and contact
If you want to discuss something, need help or found a bug, please open an issue here on Github.
The neuroformats crate was written by Tim Schäfer. You can find my email address on my website if you need to contact me.
Dependencies
~5.5MB
~99K SLoC