5 releases
| 0.1.4 | Feb 1, 2025 |
|---|---|
| 0.1.3 | Jan 31, 2025 |
| 0.1.2 | Jan 31, 2025 |
| 0.1.1 | Jan 31, 2025 |
| 0.1.0 | Jan 30, 2025 |
#240 in Machine learning
149 downloads per month
125KB
551 lines
Miniaturized Neural Evolutionary Topology
minet_ai is a minimalistic Rust library that evolves small neural networks via a genetic algorithm. Inspired by the NEAT (NeuroEvolution of Augmenting Topologies) algorithm, it focuses on producing compact, adaptive networks that grow or shrink to solve the problems they face. These networks are small, fast, and memory-efficient—ideal for NPC AI or autonomous agents where you might need hundreds or thousands of evolving neural networks running simultaneously.
Differences Between MiNET and NEAT
Fixed Hidden Neurons
A MiNET network starts with a fixed number of hidden neurons but may only use a subset of them. For example, a network with 12 hidden neurons might converge on a solution that uses only 8, leaving four neurons disconnected. This design emphasizes synapse mutations, speeding up convergence.
Gene Structure
Genes are defined per neuron (including its outgoing synapses). Every network in a population has the same number of genes (one per neuron), but each gene can vary in length depending on how many synapses it carries.
Equal Gene Swapping
When crossbreeding, MiNET networks evenly swap genes, where each child gets half its genes from each parent. Depending on the selected gene connections, this can lead to children having more or fewer total synapses than either parent.
Single Topological Mutation
During crossbreeding, a rare topological mutation removes one synapse and adds another randomly. Bias and weight mutations, however, occur every time crossbreeding occurs.
Indexed Neurons
Neurons have ascending IDs, where inputs are the lowest and outputs are the highest. Synapses can only connect to neurons with a higher ID, allowing lower-index hidden neurons to have broader connectivity and higher-index neurons to have smaller connectivity (connecting largely to output neurons). This allows fast, efficient forward passes and deeper connectivity among neurons.
Usage
Getting started and using minet_ai is extremely simple.
Create a New Network
let mut new_network = minet::new(input_neurons, hidden_neurons, output_neurons);
let mut new_network = minet::new(3, 8, 1);
Updating Fitness
Networks store their own fitness values, as an f32 where higher is better. Children of crossbreeding have zero initial fitness.
new_network.fitness += 0.1;
Running a Forward Pass
let input: Vec<f32> = vec![1.0, 2.0, 3.0];
let mut output: Vec<f32> = Vec::with_capacity(1);
let output = new_network.forward(input);
Crossbreeding
let parent_1 = minet::new(3, 8, 1);
let parent_2 = minet::new(3, 8, 1);
let child = parent_1.crossbreed(&parent_2);
Creating a Population of Networks
let network_population = minet::initialize_population(population_size, inputs, hidden, outputs);
let network_population = minet::initialize_population(100, 3, 8, 1);
Crossbreed a Population of Networks
Crossbreed a population of minet networks, retaining an elite population where surival_rate% survive and crossbreed amongst themselves to create a new population of target_population size. The survival of the elite population is based on their fitness score (minet_network.fitness).
let new_network_population = minet::crossbreed_population(network_population, survival_rate, target_population);
let new_network_population = minet::crossbreed_population(network_population, 0.1, 105);
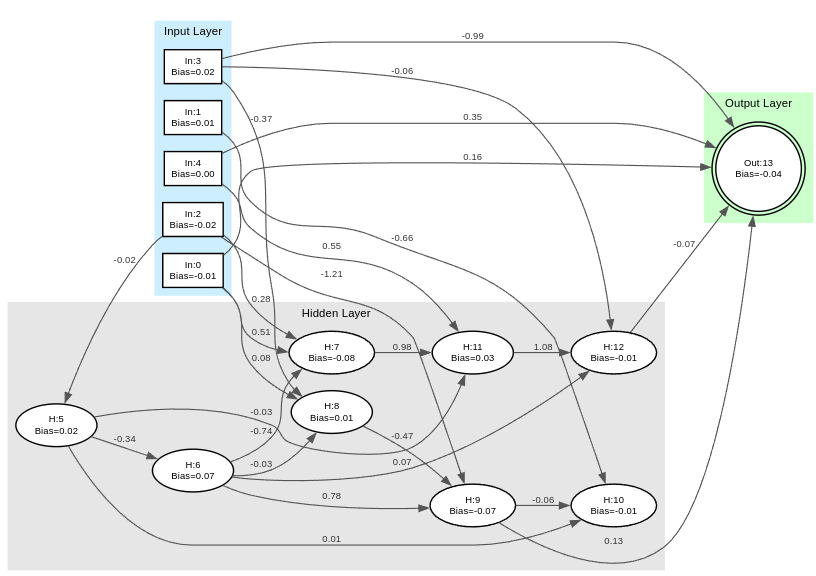
Visualize A Network as a DOT File
// Save a network to a dot file so that it can visualized with any graphviz software.
network.dot_to_file("example.dot").expect("Failed to save network to dot file");
Encode / Decode From Easily Shareable 64-Bit Representation
let network1 = minet::new(3, 5, 2);
let mut network2 = minet::new(3, 5, 2);
// Encode network_1 as a base-64 string
let network1_encoded = network1.encode();
// Import the base-64 string into network 2
network2.import_encoded(&network1_encoded);
// Now Network 1 and Network 2 are identical
// Display the genome to the console
network1.display_genome();
/*
CgAAAAAAAgNpK7Q/BK+djT8AAAAAAgVFbZw
+CbtApr0AAAAAAwO3P6+9BXLAyr4G8DxCPg
AAAAAEBBk6oj4GKtWRvgcnhnI9CBQiRj8AA
AAAAgd9zWC+CO5gJb8AAAAAAghcMIq+Ccj5
kj0AAAAAAQcvJak+AAAAAAAAAAAAAAAAAAA
*/
Display a Network to The Console
let network = minet::new(3, 5, 2);
network.display();
/*
===== GENOME =====
0 | Input :: Bias: 0, Synapses: [(3, -0.77), (4, -0.64), (5, 1.49), (7, -0.27), (8, -0.09), (9, 0.46)]
1 | Input :: Bias: 0, Synapses: [(3, -0.63), (5, -0.32), (9, 0.01)]
2 | Input :: Bias: 0, Synapses: [(3, 0.21), (4, -1.04), (8, -0.13)]
3 | Hidden :: Bias: 0, Synapses: [(4, -0.28), (5, -0.66), (6, 0.07)]
4 | Hidden :: Bias: 0, Synapses: [(6, 0.07)]
5 | Hidden :: Bias: 0, Synapses: [(7, -0.69), (8, 0.51)]
6 | Hidden :: Bias: 0, Synapses: [(7, 0.43)]
7 | Hidden :: Bias: 0, Synapses: []
8 | Output :: Bias: 0, Synapses: []
9 | Output :: Bias: 0, Synapses: []
Synapses: 19
Fitness: 0
*/
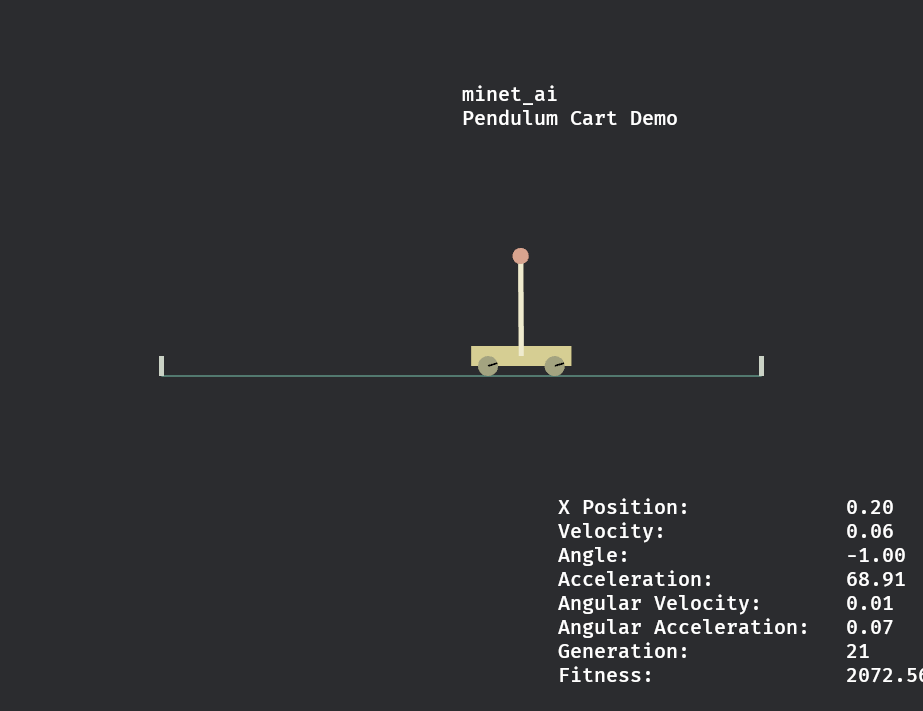
Example usage
A population of minet_ai networks learns to balance a pendulum above a cart in 21 generations.
Dependencies
~1.5MB
~25K SLoC