30 releases
| 0.5.6 | Apr 2, 2025 |
|---|---|
| 0.5.5 | Feb 10, 2025 |
| 0.5.4 | Nov 27, 2024 |
| 0.5.1 | Jul 30, 2024 |
| 0.1.1 | Nov 21, 2019 |
#17 in Biology
1,704 downloads per month
Used in 8 crates
(6 directly)
9.5MB
38K
SLoC
Bigtools 
| Rust, CLI |
|
| Python |
|
Bigtools is a library and associated tools for reading and writing bigwig and bigbed files.
The primary goals of the project are to be
- Performant
- Extensible
- Modern
Performant
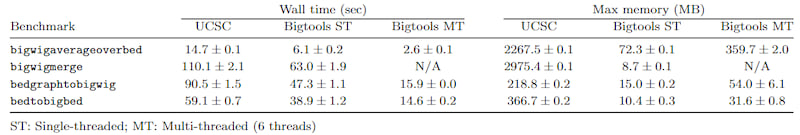
Bigtools uses async/await internally to allow for efficient, multi-core computation when possible. In addition, tools are optimized for minimal memory usage. See Benchmarks for more details.
Extensible
Bigtools is designed to be as modular as possible. This, in addition to the safety and reliability of Rust, allows both flexibility and correctness as a library. In addition, its extremely easy to quickly create new tools or binaries. A number of binaries are available that parallel related existing binaries from UCSC, with drop-in compatibility for the most common flags.
Modern
Bigtools is written in Rust and published to crates.io, so binaries can be installed with cargo install bigtools or it can be used as a library by simply including it in your cargo.toml.
Library
To use bigtools in your Rust project, add bigtools to your Cargo.toml or run:
cargo add bigtools
See the bigtools 🦀 Documentation.
Example
use bigtools::bbiread::BigWigRead;
let mut reader = BigWigRead::open("test.bigWig").unwrap();
let chr1 = reader.get_interval("chr1", 0, 10000).unwrap();
for interval in chr1 {
println!("{:?}", interval);
}
Binaries
The bigtools CLI binaries can be installed through crates.io or conda.
cargo install bigtools
conda install -c bioconda bigtools
Additionally, pre-built binaries can be downloaded through Github releases.
The following binaries are available:
| binary | description |
|---|---|
| bigtools | Provides access to multiple subcommands, including all below |
| bedgraphtobigwig | Writes a bigWig from a given bedGraph file |
| bedtobigbed | Writes a bigBed from a given bed file |
| bigbedinfo | Shows info about a provided bigBed |
| bigbedtobed | Writes a bed from the data in a bigBed |
| bigwigaverageoverbed | Calculate statistics over the regions of a bed file using values from a bigWig |
| bigwiginfo | Shows info about a provided bigWig |
| bigwigmerge | Merges multiple bigWigs, outputting to either a new bigWig or a bedGraph |
| bigwigtobedgraph | Writes a bedGraph from the data in a bigWig |
| bigwigvaluesoverbed | Get the per-base values from a bigWig over the regions of a bed file using values |
Renaming the bigtools binary to any of the subcommands (case-insensitive) allows you to run that subcommand directly.
Python wrapper
The pybigtools package is a Python wrapper written using PyO3. It can be installed or used as a dependency either through PyPI or conda.
pip install pybigtools
conda install -c bioconda pybigtools
See the pybigtools 🐍 API Documentation.
How to build from source
In order to build the bigtools binaries, you can run
cargo build --release
and the binaries can be found in target/release/.
Otherwise, you can install the binaries from source by running
cargo install --path bigtools/
Building the python wheels for pybigtools requires maturin. To build the pybigtools wheel for installation (and install), you can run
maturin build --release -m pybigtools/Cargo.toml
pip install target/wheels/pybigtools*.whl
or
maturin develop --release -m pybigtools/Cargo.toml
Benchmarks
Benchmarks are included in the ./bench directory. They require python to run.
Multiple tools are compared against the comparable UCSC tools. For completeness, both single-threaded and multi-threaded (when available) benchmarks are included. Multiple different configuration options are benchmarked across multiple replicates, but a summar is available in the table below:
How to cite
This repository contains contains a CITATION.cff file with citation information. Github allows you to get a citation in either APA or BibTeX format; this is available in "Cite this repository" under About.
Dependencies
~3–15MB
~193K SLoC