4 releases (2 breaking)
| 0.3.0 | Jun 10, 2024 |
|---|---|
| 0.2.0 | Mar 12, 2024 |
| 0.1.1 | Mar 7, 2024 |
| 0.1.0 | Jan 22, 2024 |
#766 in Machine learning
1.5MB
5K
SLoC
What's in my big data?
This repository contains the code for running What's In My Big Data (WIMBD), which accompanies our recent paper (with the same name).
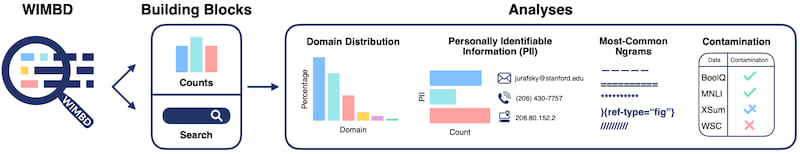
What is WIMBD?
WIMBD is composed of two components
- A set of tools for analyzing and revealing the content of large-scale datasets
- A set of analyses we apply to those datasets, using the aforementioned tools
WIMBD tools consist of two parts:
- Count
- Search
The count follows a map-reduce functionality, which divides the task into smaller chunks, applies the operation (e.g., extract the domain from a URL) and then aggregates the counts. We have two implementations for this. One through python functions (e.g., for domain counts) which is easily extendable and scalable, and one through a Rust CLI for faster processing. The Rust implementation covers the summary statistics (presented in Table 2 in the paper) such as the corpus size, number of tokens, etc. In addition, it computes the most & least common $n$-grams approximation using counting Bloom filters.
In practice, we implement search using elasticsearch. We index 5 of the corpora we consider, and provide both a UI and a programmatic access to those.
We built some wrappers around the ES API, which allows count and extract functionalities. We provide a more detailed documentation here.
Getting started
There are two distinct parts of this toolkit: a Python library of functions and a Rust-based CLI.
Using the Python library
Create python environment
conda create -n wimbd python=3.9
conda activate wimbd
pip install -r requirements.txt
export PYTHONPATH="${PYTHONPATH}:/PATH/TO/wimbd/"
As an example, run the following command that counts the domain counts, per token (Section 4.2.2 in the paper):
bash wimbd/url_per_tok_counts/run.sh /PATH-TO/c4/en/c4-train.* > data/benchmark/benchmark_url_tok_c4.jsonl
Run scheme counts
./wimbd/scheme_counts/run.sh /PATH-TO/laion2B-en/*.gz > data/scheme_laion2B-en.jsonl
This will run the map reduce scripts, and dump the results into a file
Using the Rust CLI
This part of the repository is written in Rust, so first you'll have to install the Rust toolchain. There's a simple one-liner for that:
curl --proto '=https' --tlsv1.2 -sSf https://sh.rustup.rs | sh
Then you can either install the latest release from crates.io directly or install from source.
To install from crates.io, run:
cargo install wimbd
Or to install from source, run:
make release DIR=./bin
(make sure to change DIR to a directory of your choice that's on your PATH)
And now you should have be able to run the wimbd CLI:
wimbd --help
For example, find the top 20 3-grams in some c4 files with:
wimbd topk \
/PATH-TO/c4/en/c4-train.01009-of-01024.json.gz \
/PATH-TO/c4/en/c4-train.01010-of-01024.json.gz \
-n 3 \
-k 20 \
--size 16GiB
Search
Due to the nature of ElasticSearch, we cannot release the API keys on the web. If you are interested in using our ElasticSearch indices, please fill up this form, and we'll get back to you as soon as we can.
Issues
If there's an issue with the code, or you have questions, feel free to open an issue or send a PR
Dependencies
~19–29MB
~477K SLoC