4 releases (2 breaking)
Uses old Rust 2015
| 0.3.0 | Aug 23, 2019 |
|---|---|
| 0.2.1 | Feb 21, 2018 |
| 0.2.0 | Feb 21, 2018 |
| 0.1.0 | Nov 9, 2017 |
#387 in Machine learning
77KB
871 lines
random-world 

This is a rust implementation of Machine Learning (ML) methods for confident
prediction (e.g., Conformal Predictors) and related ones introduced in the book
Algorithmic Learning in a Random World (ALRW),
which also provides standalone binaries.
Goals
These are the main goals of this library.
- Fast implementation of methods described in ALRW book
- Be well documented
- Provide standalone binaries
- CP method should be able to use scoring classifiers from existing ML libraries (e.g., rusty-machine, rustlearn)
- Easily interface with other languages (e.g., Python)
Binaries
Standalone binaries are meant to cover most functionalities of the library.
They operate on .csv files, and allow to make CP predictions, test exchangeability,
and much more.
Installation
To install the binaries, install Rust's package manager cargo. Then run:
$ cargo install random-world
This will install on your system the binaries: cp-predict, icp-predict,
and martingales.
cp-predict
cp-predict allows making batch and on-line CP predictions.
It runs CP on a training set, and uses it to predict a test set;
each dataset should be contained in a CSV file with rows:
label, x1, x2, ...
where label is a label id, and x1, x2, ...
are the values forming a feature vector.
It is important that label ids are 0, 1, ..., n_labels-1 (i.e., with no
missing values); one can specify --n-labels if not all labels
are available in the initial training data (e.g., in on-line mode).
Results are returned in a CSV file with rows:
p1, p2, ...
where each value is either a prediction (true/false) or
a p-value (float in [0,1]), depending on the flags passed to cp-predict;
each row contains a value for each label.
Example:
$ cp-predict knn -k 1 predictions.csv train_data.csv test_data.csv
Runs CP with nonconformity measure k-NN (k=1) on train_data.csv,
predicts test_data.csv, and stores the output into
predictions.csv.
The default output are p-values; to output actual predictions, specify
a significance level with --epsilon.
To run CP in on-line mode on a dataset (i.e., predict one object per time and then append it to the training examples), only specify the training file:
$ cp-predict knn -k 1 predictions.csv train_data.csv
More options are documented in the help:
$ cp-predict -h
Predict data using Conformal Prediction.
If no <testing-file> is specified, on-line mode is assumed.
Usage: cp knn [--knn=<k>] [options] [--] <output-file> <training-file> [<testing-file>]
cp kde [--kernel<kernel>] [--bandwidth=<bw>] [options] [--] <output-file> <training-file> [<testing-file>]
cp (--help | --version)
Options:
-e, --epsilon=<epsilon> Significance level. If specified, the output are
label predictions rather than p-values.
-s, --smooth Smooth CP.
--seed=<s> PRNG seed. Only used if --smooth set.
-k, --knn=<kn> Number of neighbors for k-NN [default: 5].
--n-labels=<n> Number of labels. If specified in advance it
slightly improves performances.
-h, --help Show help.
--version Show the version.
icp-predict
The syntax for icp-predict is currently identical to that of cp-predict:
the size of the calibration set is chosen to be half the size of the training
set.
This will change (hopefully soon).
martingales
Computes exchangeability martingales from a file of p-values.
P-values should be computed using cp-predict in an on-line setting
for a single label problem.
$ martingales -h
Test exchangeability using martingales.
Usage: martingales plugin [--bandwidth=<bw>] [options] <output-file> <pvalues-file>
martingales power [--epsilon=<e>] [options] <output-file> <pvalues-file>
martingales (--help | --version)
Options:
--seed PRNG seed.
-h, --help Show help.
--version Show the version.
Example:
Have a look at examples/non-iid.csv:
0,1.77750,-0.84078
0,-1.68787,3.86305
0,-0.56455,0.17416
0,-0.86380,0.39916
...
These represent anomalous time series data, artificially generated to serve as an
example for this very code; do not use it as an anomaly detection benchmark:
it will have little use.
Each line contains a label (always set to 0, which is required for computing martingales in
the next steps), and a vector. These data contains 200 examples: the first
100 were generated according to a multivariate Normal distribution centered
in [0, 0] and with covariance matrix [[1, 0], [0, 5]]; the last
200 examples according to a multivariate Normal distribution with mean
[5, 5] and covariance [[5, 0], [0, 10]].
Therefore, the "anomalous" section of the data begins after 100 examples.
To produce the martingales:
# Use CP in on-line mode
cp-predict knn -k 1 pvalues.csv examples/non-iid.csv
# Martingales
martingales plugin --bandwidth=0.2 martingales.csv pvalues.csv
Have a look at martingales.csv: it should now contain the martingale
values, one per line.
You can call anomalous behaviour whenever the
martingale's value is over a threshold (e.g., 20 or 100).
Here's a plot of the martingales values for this specific example.
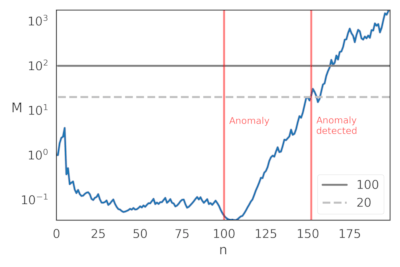
With threshold 20, the anomaly is detected roughly 50 examples after the anomaly happens.
Library
To exploit all the functionalities, or to integrate it into your project, you may want to use the actual library.
Include the following in Cargo.toml:
[dependencies]
random-world = "0.2.1"
Quick Intro
Using a deterministic (i.e., non smooth) Conformal Predictor with k-NN
nonconformity measure (k=2) and significance level epsilon=0.3.
The prediction region will contain the correct label with probability
1-epsilon.
#[macro_use(array)]
extern crate ndarray;
extern crate random_world;
use random_world::cp::*;
use random_world::ncm::*;
// Create a k-NN nonconformity measure (k=2)
let ncm = KNN::new(2);
// Create a Conformal Predictor with the chosen nonconformity
// measure and significance level 0.3.
let mut cp = CP::new(ncm, Some(0.3));
// Create a dataset
let train_inputs = array![[0., 0.],
[1., 0.],
[0., 1.],
[1., 1.],
[2., 2.],
[1., 2.]];
let train_targets = array![0, 0, 0, 1, 1, 1];
let test_inputs = array![[2., 1.],
[2., 2.]];
// Train and predict
cp.train(&train_inputs.view(), &train_targets.view())
.expect("Failed prediction");
let preds = cp.predict(&test_inputs.view())
.expect("Failed to predict");
assert!(preds == array![[false, true],
[false, true]]);
Please, read the docs for more examples.
Features
Methods:
- Deterministic and smoothed Conformal Predictors (aka, transductive CP)
- Deterministic Inductive Conformal Predictors (ICP)
- Plug-in and Power martingales for exchangeability testing
- Venn Predictors
Nonconformity measures:
- k-NN
- KDE
- Generic wrapper around ML scorers from existing libraries (e.g., rusty-machine)
Binaries:
- CP (both batch prediction and on-line)
- Martingales
- Inductive CP (batch prediction only)
Bindings:
- Python bindings
Authors
- Giovanni Cherubin (giocher.com)
Similar Projects
- nonconformist is a Python implementation of CP and ICP.
Dependencies
~10MB
~192K SLoC