3 stable releases
| 1.1.1 | Jan 30, 2023 |
|---|---|
| 1.1.0 | Nov 25, 2022 |
| 1.0.0 | Oct 14, 2022 |
| 1.0.0-alpha.1 |
|
| 0.0.0 |
|
#158 in Biology
23 downloads per month
165KB
1K
SLoC
INTRODUCING NUCLEOB
nucleob is a tool that performs statistical analysis on .fasta files containing nucleotide or amino acid sequences. It is engineered to produce highly reliable, clearly formatted results while maintaining a minimalistic design and cross-platform compatibility.
[keywords: amino acids, bioinformatics, biology, DNA, fasta, genes, genetics, molecular, nucleic acids, nucleobases, nucleotides, proteins, RNA, scientific computing, statistics, stats]
EXAMPLES
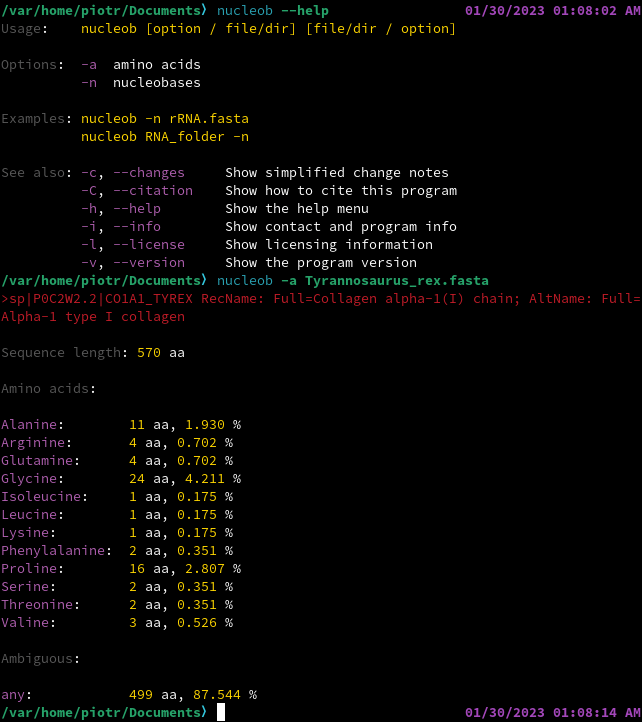
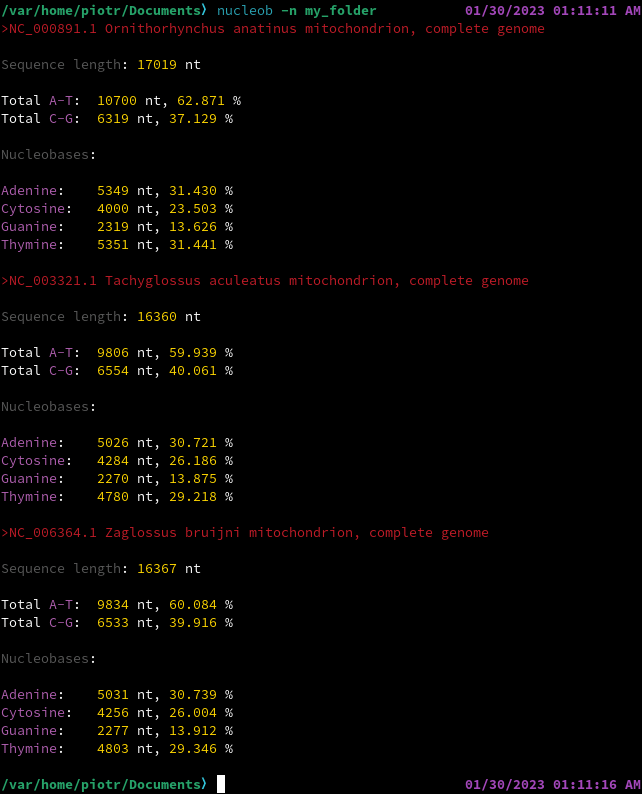
CITATION AND REUSE
nucleob can be modified and forked under the terms of the MIT License. When citing nucleob in research articles, it is important to refer to a specific program version, such as:
Bajdek, P., 2023. nucleob (version 1.1.1). [computer software] https://github.com/piotrbajdek/nucleob
INSTALLATION ON LINUX
nucleob is designed to be compatible with Windows and macOS, and can be easily installed using cargo. However, the primary development and testing environment for nucleob is Fedora Linux.
The current version of nucleob (v1.1.1) has been verified to work properly on Fedora Linux 37 and Ubuntu 22.10.
METHOD 1 – USING CARGO
[Recommended for programmers]
1. To install nucleob from crates.io, use the following cargo command:
cargo install nucleob
The executable will be saved in the hidden .cargo/bin/ directory within your home directory.
2a. For easy access, you may want to copy the nucleob file to the /usr/bin/ directory. This can be done by following the instructions in Method 2 (3a, 3b).
2b. As an alternative, you can add the ~/.cargo/bin/ directory to your system's PATH variable, which can be configured using rustup.
METHOD 2 – UNIVERSAL LINUX BINARIES
1. To install nucleob, first download the distro-independent binary from GitHub.
2. Then, make the file executable by running the command:
sudo chmod +x ./nucleob
3a. On most Linux distributions, install nucleob by copying the binary to /usr/bin/:
sudo cp nucleob /usr/bin/
3b. For Fedora Silverblue / Kinoite, use this command:
sudo cp nucleob /var/usrlocal/bin/
METHOD 3 – DISTRO-SPECIFIC PACKAGES
[Recommended for most users]
Distro-specific packages for .rpm and .deb-based Linux distributions are also available for download. To install nucleob on different Linux distributions, follow these instructions:
Fedora Linux / RHEL / openSUSE:
sudo rpm -i nucleob-1.1.1-1.x86_64.rpm
Fedora Silverblue / Kinoite:
rpm-ostree install nucleob-1.1.1-1.x86_64.rpm
Ubuntu:
sudo dpkg -i nucleob_1.1.1_amd64.deb
METHOD 4 – MANUAL COMPILATION
First, download and unpack the nucleob source code from GitHub. Next, to build and install the program, use the command:
cargo build --release && sudo cp target/release/nucleob /usr/bin/