5 releases
| 0.1.4 | Sep 14, 2024 |
|---|---|
| 0.1.3 | Sep 14, 2024 |
| 0.1.2 | Sep 13, 2024 |
| 0.1.1 | Sep 6, 2024 |
| 0.1.0 | Sep 6, 2024 |
#656 in Text processing
540KB
1.5K
SLoC
Abbreviation Extractor
Abbreviation Extractor is a high-performance Rust library with Python bindings for extracting abbreviation-definition pairs from text, particularly focused on biomedical text. It implements an improved version of the Schwartz-Hearst algorithm, offering enhanced accuracy and speed. It's based the original python implementation.
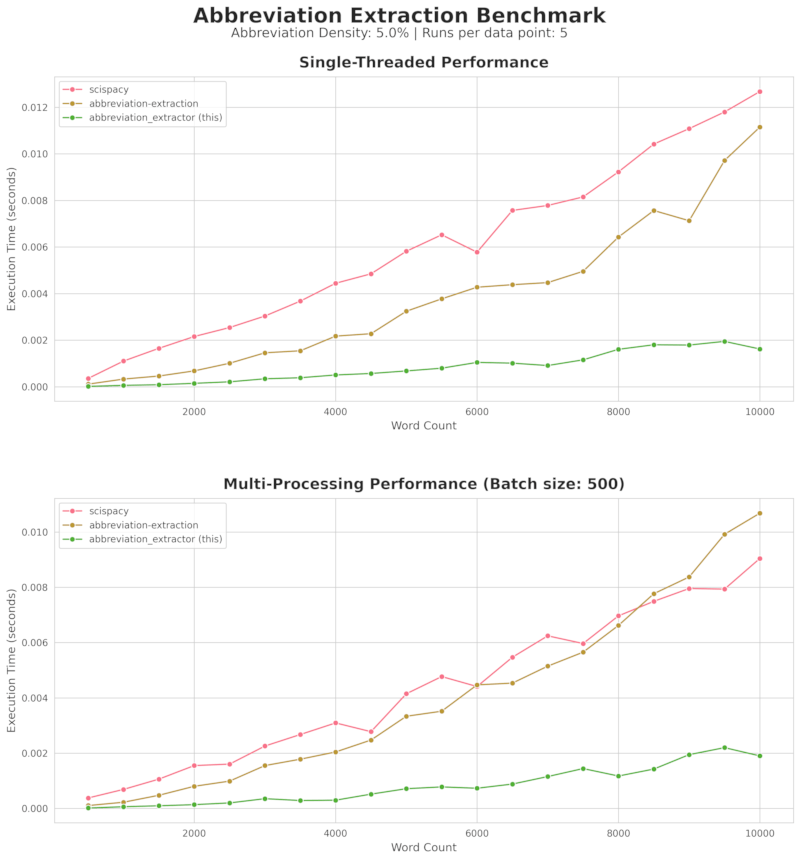
Speed Comparison With Other Abbreviation Extraction Libraries
Extraction Accuracy Comparison With Other Abbreviation Extraction Libraries
Features
- Fast and accurate extraction of abbreviation-definition pairs with tokenization.
- Support for both single-threaded and parallel processing
- Python bindings for easy integration with Python projects
- Customizable extraction parameters like selecting the most common or first definition for each abbreviation
Installation
Rust
Add this to your Cargo.toml:
abbreviation-extractor = "0.1.3"
Python
pip install abbreviation-extractor-rs
Basic Usage
Rust
use abbreviation_extractor::{extract_abbreviation_definition_pairs, AbbreviationOptions};
let text = "The World Health Organization (WHO) is a specialized agency.";
let options = AbbreviationOptions::default();
let result = extract_abbreviation_definition_pairs(text, options);
for pair in result {
println!("Abbreviation: {}, Definition: {}", pair.abbreviation, pair.definition);
}
Python
from abbreviation_extractor import extract_abbreviation_definition_pairs
text = "The World Health Organization (WHO) is a specialized agency."
result = extract_abbreviation_definition_pairs(text)
for pair in result:
print(f"Abbreviation: {pair.abbreviation}, Definition: {pair.definition}")
Customizing Extraction
Python
from abbreviation_extractor import extract_abbreviation_definition_pairs
text = "The World Health Organization (WHO) is a specialized agency. The World Heritage Organization (WHO) is different."
# Get only the most common definition for each abbreviation
result = extract_abbreviation_definition_pairs(text, most_common_definition=True)
# Get only the first definition for each abbreviation
result = extract_abbreviation_definition_pairs(text, first_definition=True)
# Disable tokenization (if the input is already tokenized)
result = extract_abbreviation_definition_pairs(text, tokenize=False)
# Combine options
result = extract_abbreviation_definition_pairs(text, most_common_definition=True, tokenize=True)
for pair in result:
print(f"Abbreviation: {pair.abbreviation}, Definition: {pair.definition}")
Rust
use abbreviation_extractor::{extract_abbreviation_definition_pairs, AbbreviationOptions};
let text = "The World Health Organization (WHO) is a specialized agency. The World Heritage Organization (WHO) is different.";
// Get only the most common definition for each abbreviation
let options = AbbreviationOptions::new(true, false, true);
let result = extract_abbreviation_definition_pairs(text, options);
// Get only the first definition for each abbreviation
let options = AbbreviationOptions::new(false, true, true);
let result = extract_abbreviation_definition_pairs(text, options);
// Disable tokenization (if the input is already tokenized)
let options = AbbreviationOptions::new(false, false, false);
let result = extract_abbreviation_definition_pairs(text, options);
for pair in result {
println!("Abbreviation: {}, Definition: {}", pair.abbreviation, pair.definition);
}
Parallel Processing
For processing multiple texts in parallel, you can use the extract_abbreviation_definition_pairs_parallel function:
Rust
use abbreviation_extractor::{extract_abbreviation_definition_pairs_parallel, AbbreviationOptions};
let texts = vec![
"The World Health Organization (WHO) is a specialized agency.",
"The United Nations (UN) works closely with WHO.",
"The European Union (EU) is a political and economic union.",
];
let options = AbbreviationOptions::default();
let result = extract_abbreviation_definition_pairs_parallel(texts, options);
for extraction in result.extractions {
println!("Abbreviation: {}, Definition: {}", extraction.abbreviation, extraction.definition);
}
Python
from abbreviation_extractor import extract_abbreviation_definition_pairs_parallel
texts = [
"The World Health Organization (WHO) is a specialized agency.",
"The United Nations (UN) works closely with WHO.",
"The European Union (EU) is a political and economic union.",
]
result = extract_abbreviation_definition_pairs_parallel(texts)
for extraction in result.extractions:
print(f"Abbreviation: {extraction.abbreviation}, Definition: {extraction.definition}")
Processing Large Files
For extracting abbreviations from large files, you can use the extract_abbreviations_from_file function:
Rust
use abbreviation_extractor::{extract_abbreviations_from_file, AbbreviationOptions, FileExtractionOptions};
let file_path = "path/to/your/large/file.txt";
let abbreviation_options = AbbreviationOptions::default();
let file_options = FileExtractionOptions::default();
let result = extract_abbreviations_from_file(file_path, abbreviation_options, file_options);
for extraction in result.extractions {
println!("Abbreviation: {}, Definition: {}", extraction.abbreviation, extraction.definition);
}
Python
from abbreviation_extractor import extract_abbreviations_from_file
file_path = "path/to/your/large/file.txt"
result = extract_abbreviations_from_file(file_path)
for extraction in result.extractions:
print(f"Abbreviation: {extraction.abbreviation}, Definition: {extraction.definition}")
You can customize the file extraction process by specifying additional parameters:
result = extract_abbreviations_from_file(
file_path,
most_common_definition=True,
first_definition=False,
tokenize=True,
num_threads=4,
show_progress=True,
chunk_size=2048 * 1024 # 2MB chunks
)
Benchmark
Below is a comparison of how the abbreviation extractor performs in comparison to other libraries, namely Schwartz-Hearst and ScispaCy in terms of accuracy and speed.
Performance Comparison of Abbreviation Extractor Against Other Libraries
| Abbrv | Ground Truth | abbreviation-extractor (This Library) | abbreviation-extraction | ScispaCy |
|---|---|---|---|---|
| '3-meAde' | '3-methyl-adenine' | '3-methyl-adenine' | '3-methyl-adenine' | 'N/A' |
| '5'UTR' | '5' untranslated region' | '5' untranslated region' | 'N/A' | 'N/A' |
| '5LO' | '5-lipoxygenase' | '5-lipoxygenase' | '5-lipoxygenase' | 'N/A' |
| 'AAV' | 'adeno-associated virus' | 'adeno-associated virus' | 'associated virus' | 'adeno-associated virus' |
| 'ACP' | 'Enoyl-acyl carrier protein' | 'Enoyl-acyl carrier protein' | 'acyl carrier protein' | 'Enoyl-acyl carrier protein' |
| 'ADIOL' | '5-androstene-3beta, 17beta-diol' | '5-androstene-3beta, 17beta-diol' | 'androstene-3beta, 17beta-diol' | '5-androstene-3beta, 17beta-diol' |
| cAMP | 'cyclic AMP' | 'cyclic AMP' | 'N/A' | |
| 'ALAD' | '5-aminolaevulinic acid dehydratase' | '5-aminolaevulinic acid dehydratase' | 'N/A' | '5-aminolaevulinic acid dehydratase' |
| 'AMPK' | 'AMP-activated protein kinase' | 'AMP-activated protein kinase' | 'N/A' | 'AMP-activated protein kinase' |
| 'AP' | 'apurinic/apyrimidinic site' | 'apurinic/apyrimidinic site' | 'apyrimidinic site' | 'apurinic/apyrimidinic site' |
| 'AcCoA' | 'acetyl coenzyme A' | 'acetyl coenzyme A' | 'N/A' | 'acetyl coenzyme A' |
| 'Ahr' | 'aryl hydrocarbon receptor' | 'aryl hydrocarbon receptor' | 'N/A' | 'aryl hydrocarbon receptor' |
| 'BD' | 'binding domain' | 'binding domain' | 'N/A' | 'binding domain' |
| '8-OxoG' | '7,8-dihydro-8-oxoguanine' | '7,8-dihydro-8-oxoguanine' | '8-oxoguanine' | 'N/A' |
| dsRNA | double-stranded RNA | double-stranded RNA | double-stranded RNA | 'N/A' |
| 'BERI' | 'Biomolecular Engineering Research Institute' | 'Biomolecular Engineering Research Institute' | 'N/A' | 'Biomolecular Engineering Research Institute' |
| 'CTLs | 'cytotoxic T lymphocytes' | 'cytotoxic T lymphocytes' | 'N/A' | 'N/A' |
| 'C-RBD' | 'C-terminal RNA binding domain' | 'C-terminal RNA binding domain' | 'N/A' | 'C-terminal RNA binding domain' |
| 'CAP' | 'cyclase-associated protein' | 'cyclase-associated protein' | 'N/A' | 'cyclase-associated protein' |
Speed Comparison with Other Abbreviation Extraction Libraries
API Reference
For detailed API documentation, please refer to the Rust docs or the Python module docstrings.
Contributing
Contributions are welcome! Please feel free to submit a Pull Request.
License
This project is licensed under the MIT License.
Acknowledgements
This library is based on the Schwartz-Hearst algorithm:
The implementation is inspired by the original Python variant by Phil Gooch: abbreviation-extractor
Dependencies
~16–25MB
~261K SLoC