22 releases (8 breaking)
| 0.9.2 | Nov 18, 2024 |
|---|---|
| 0.8.3 | Sep 22, 2024 |
| 0.7.4 | Jul 15, 2024 |
| 0.6.1 | Mar 28, 2024 |
| 0.1.1 | Mar 27, 2023 |
#207 in Parser implementations
Used in 2 crates
325KB
6.5K
SLoC

STAM Tools
A collection of command-line tools for working with STAM, a data-model for stand-off annotations on text.
Various tools are grouped under the stam tool, and invoked with a subcommand:
stam align- Align two similar texts, mapping their coordinate spaces.stam annotateorstam add- Add annotations or datasets or resources (from file or by query).stam batchorstam shell- Process multiple subcommands in sequence, or run interactively.stam info- Return information regarding a STAM model.stam init- Initialize a new STAM annotationstore (either from scratch or as a copy/merge of others)stam import- Import STAM data in tabular from a simple TSV (Tab Separated Values) format, allows custom columns.stam fromxml- Import data from XML-based formats (like xHTML, TEI) to STAM. Effectively 'untangling' text and annotations.stam print- Output the text of any resources in the model.stam queryorstam export- Query the annotation store and export the output in tabular form to a simple TSV (Tab Separated Values) format. This is not lossless but provides a decent view on the data. It provides a lot of flexibility by allowing you to configure the output columns as you see fit.stam validate- Validate a STAM model.stam tag- Regular-expression based tagger on plain text.stam split- Split an annotation store by removing specific resources, data sets or annotations.stam view- View annotations as queried by outputting to HTML (or ANSI coloured text).
For many of these, you can set --verbose for extra details in the output.
Alternatively, the functionality provided by the tools is also exposed as a library via a Rust API.
Installation
From source
$ cargo install stam-tools
Demo
Usage
Add the --help flag after the subcommand for extensive usage instructions.
Most tools take as input a STAM JSON or CSV file containing an annotation store. You
may also specify multiple stores which will be merged into one. Any files
mentioned via the @include mechanism are loaded automatically.
When output is written, the first store file used as input is also used as
output. You can prevent writing output files by setting --dry-run or prevent
reusing the first input file by setting an explicit output using --output.
Instead of passing STAM JSON files, you can read from stdin and/or output to
stdout by setting the filename to -, this works in many places.
These tools also support reading and writing STAM CSV.
Tools
stam init & stam annotate
The stam init command is used to initialize a new STAM annotationstore with
resources (--resource, plain text or STAM JSON), annotation data sets
(--annotationset, STAM JSON) and/or annotations (--annotations, JSON list
of annotations in STAM JSON).
Example, the positional parameter (last one) is the annotation store to output, it may be STAM JSON or STAM CSV, determined by the file extension:
$ stam init --resource document.txt new.store.stam.json
The stam annotate command is almost identical to stam init, except it reads
and modifies an existing annotation store, rather than starting a new one from
scratch:
$ stam annotate --resource document.txt existing.store.stam.json
Whenever you load annotations and annotation data sets using these commands,
they need to already be in STAM JSON format. To import data from other formats,
use stam import instead.
The stam init and stam annotate commands are also capable of merging
multiple annotation stores into one.
If you want to load a STAM annotationstore (or multiple) and save it under
another name and/or other format, you can use stam init (or stam annotate)
as well, they key is to then use an explicit --output filename that differs
from the input. It serves to merge stores and/or convert between STAM JSON and
STAM CSV. Example:
$ stam init --output merged.store.stam.csv mystore1.store.stam.json mystore2.store.stam.json
You can also pass STAMQL queries
to stam annotate to add (or delete) annotations:
stam annotate --query 'ADD ANNOTATION WITH DATA "my-vocab" "type" "sentence"; TARGET ?x { SELECT TEXT ?x WHERE RESOURCE "smallquote.txt" OFFSET 0 25; }' demo.store.stam.json
stam info
The stam info command provides either some high-level details on the
annotation store (number of resource, annotations, etc), or with the
--verbose flag it goes as far as presenting, in a fairly raw format, all the
data it holds.
Example:
$ stam info my.store.stam.json
stam query
The stam query tool is used to consult the annotation store and export
selected STAM data into a simple tabular data format (TSV, tab separated
values). You can configure precisely what columns you want to export using the
--columns parameter, or simply rely on the defaults that are autodetected.
See stam query --help for a list of supported columns.
A full query is done using the --query parameter and subsequently a query
statement in the STAM Query Language
(STAMQL):
Example 1) a query in STAMQL:
$ stam query --query 'SELECT ANNOTATION ?a WHERE DATA "myset" "pos" = "noun";'
However, if you simply want all annotations, resource, data, and don't want to formulate a query a shortcut is
available by just the --type parameter to annotation,key,data,resource or dataset.
Example 2) get all annotations (also default behaviour if you omit --type and --query):
$ stam query --type annotation my.store.stam.json
For certain types, you can set --verbose to output more information, e.g.
when querying for annotations it will also output all annotation data
pertaining to the annotations. Do not that stam import can not import
annotations back when you use this.
Example 3) get all annotations verbosely with all data:
$ stam query --verbose --type annotation my.store.stam.json
Example 4) get all keys:
$ stam query --type key my.store.stam.json
One of the more powerful functions is that you can specify custom columns by
specifying a set ID, a delimiter and a key ID (the delimiter by default is a
slash), for instance: my_set/part_of_speech. Such columns are automatically
added for you if you have DATA or KEY constraints in your query (like in
example 1), if that is not what you want, set --strict-columns. This custom column will hold
the corresponding value if they key exists for the annotation.
Example 5) explicitly specified columns including a custom one:
$ stam query --columns Id,Text,TextResource,BeginOffset,EndOffset,my_set/part_of_speech my.store.stam.json
Example 6) Subqueries and multiple result variables
$ stam query --query 'SELECT ANNOTATION ?sentence WHERE DATA "myset" "type" = "sentence"; { SELECT ANNOTATION ?word WHERE RELATION ?sentence EMBEDS; DATA "myset" "type" = "word"; }'
This will result in a TSV file where the sentence will be repeated for each word that is found in it, a result number will be returned in a column, as well as the variable name.
The TSV output produced by this tool is not lossless, that is, it can not encode everything that STAM supports, unlike STAM JSON and STAM CSV. It does, however, give you a great deal of flexibility to quickly output only the data relevant for whatever your specific purpose is.
For queries that modify the annotation store, use stam annotate rather than stam query.
stam export
stam export is just an alias for stam query, their functionality is identical.
stam import
The stam import tool is used to import tabular data from a TSV (Tab Separated
Values) file into STAM. Like stam query, you can configure precisely what
columns you want to import, using the --columns parameter. By default, the
import function will attempt to parse the first line of your TSV file as the
header and use that to figure out the column configuration. You will often
want to set --annotationset to set a default annotation set to use for
custom columns. If you set --annotationset my_set then a column like
part_of_speech will be interpreted in that set (same as if you wrote
my_set/part_of_speech explicitly).
Here is a simple example of a possible import TSV file (with --annotationset my_set):
Text TextResource BeginOffset EndOffset part_of_speech
Hello hello.txt 0 5 interjection
world hello.txt 6 10 noun
The import function has some special abilities. If your TSV data does not
mention specific offsets in a text resource(s), they will be looked up
automatically during the import procedure! If the text resources don't even
exist in the first place, they can be reconstructed (within certain
constraints, the output text will likely be in tokenised form only). If your
data does not explicitly reference a resource, use the --resource parameter
to point to an existing resource that will act as a default, or
--new-resource for the reconstruction behaviour.
By setting --resource hello.txt or --new-resource hello.txt you can import the following much more minimal TSV:
Text part_of_speech
Hello interjection
world noun
The importer supports empty lines within the TSV file. When reconstructing
text, these will map to (typically) a newline in the to-be-constructed text
(this configurable with --outputdelimiter2). Likewise, the delimiter
between rows is configurable with --outputdelimiter, and defaults to a space.
Note that stam import can not import everything stam query can export. It can only import rows
exported with --type Annotation (the default), in which each row
corresponds with one annotation.
stam grep
The stam grep tool can be used for matching regular expressions in text,
it will return the resource identifiers, offsets and exact texts of all matching occurrences.
Example:
$ stam grep -e "[hzwHZW]ij" frogdeep.store.stam.json
example.deep 690:693 Hij 1/1
example.deep 799:802 hij 1/1
The tab-separated columns are as follows:
- Resource ID
- Begin offset and end offset (non-inclusive) in unicode points
- The matching text
- The current capture group and total number of capture groups (if any)
stam tag
The stam tag tool can be used for matching regular expressions in text and
subsequently associating annotations with the found results. It is a tool to do
for example tokenization or other tagging tasks.
The stam tag command takes a TSV file (example) containing regular expression rules for the tagger.
The file contains the following columns:
- The regular expressions follow the this syntax. The expression may contain one or or more capture groups containing the items that will be tagged, in that case anything else is considered context and will not be tagged.
- The ID of annotation data set
- The ID of the data key
- The value to set. If this follows the syntax $1,$2,etc.. it will assign the value of that capture group (1-indexed).
Example of the rules:
#EXPRESSION #ANNOTATIONSET #DATAKEY #DATAVALUE
\w+(?:[-_]\w+)* simpletokens type word
[\.\?,/]+ simpletokens type punctuation
[0-9]+(?:[,\.][0-9]+) simpletokens type number
Example of applying this to a text resource:
# first we create a store and add a text resource
$ stam init --resource sometext.txt my.store.stam.json
# then we start the tagging
$ stam tag --rules rules.tsv my.store.stam.json
stam view
The stam view tool is used to visualize annotations. The default
visualisation is HTML. This will output a self-contained static HTML document
to standard output (the document does not reference any external assets). An
alternative visualisation is text with ANSI escape codes for colours (--format ansi), which is suited for display in a terminal rather than a browser. The
annotations you want to visualise are requested via queries in
STAMQL,
using the --query parameter.
The first query you have may contains subqueries which act as highlight queries. The main query is always the selection query, it determines what the main selection is and can be anything you can query that has text (i.e. resources, annotations, text selections).
Any subsequent queries are highlight queries, they determine what parts of the selections produced by the selection query you want to highlight. Highlighting is done by drawing a line underneath the text and optionally by a tag that shows extra information.
Instead of specifying subqueries, you may use the --query parameter multiple
times to define subqueries via the command line. Always make sure these
reference a variable defines in the main query.

Example with tags:
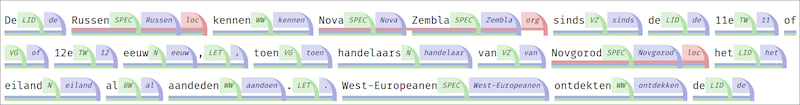
Tags can be enabled by prepending the query/subquery (i.e. before SELECT) with one of the following attributes:
@KEYTAG- Outputs a tag with the key@KEYVALUETAG- Outputs a tag with the key and the value@VALUETAG- Outputs a tag with the value only@IDTAG- Outputs a tag with the public identifier of the ANNOTATION that has been selected
The first three tags use to the first DATA constraint found, if put before
SELECT. Alternatively, they can also be directly put before (any) DATA
constraint to explicitly select one.
Attributes may also be provided for styling HTML output, these go before the query/subquery as a whole:
@STYLE=class - Will associate the mentioned CSS class (it's up to you to associate a proper stylesheet). The default one predefines only a few simple classes:italic,bold,red,green,blue,super.@HIDE- Do not add the highlight underline and do not add an entry to the legend. This may be useful if you only want to apply@STYLE.
If no attribute is provided, there will be no tags or styling shown for that query, only a highlight underline.
In the highlight queries, the variable from the main
selection query is available and you should always use it in a constraint, otherwise
performance will be sub-optimal! All your queries should have variable names
and these will appear in the legend (unless you pass --no-legend).
Various real examples of visualisation and queries are shown here: https://github.com/knaw-huc/stam-experiments/tree/main/exp6
Example of ANSI output rather than HTML, using --format ansi:
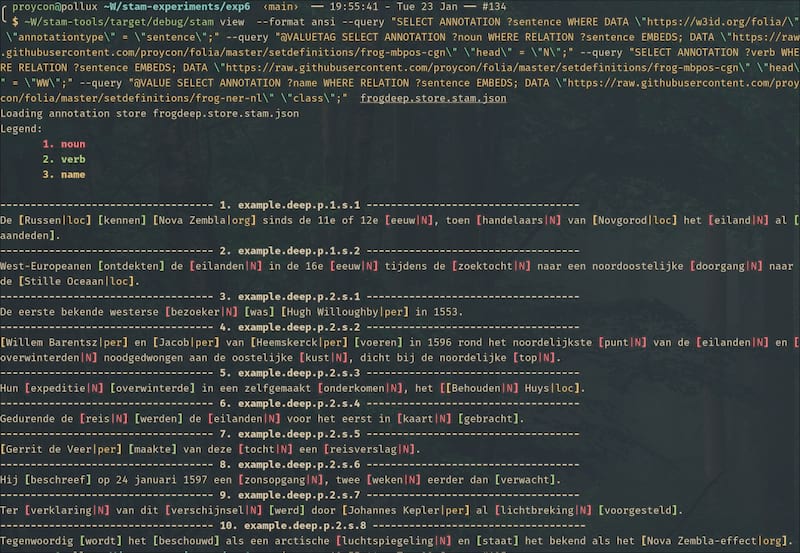
stam align
The stam align tool is used to compute an alignment between two texts; it
identifies which parts of the two texts are identical and computes a mapping
between the two coordinate systems. Two related sequence alignments algorithms
from bioinformatics are implemented to accomplish this:
Smith-Waterman
and Needleman-Wunsch.
The score parameters to either are fully configurable.
The resulting alignment is added as an annotation, a so called transposition, according to the STAM Transpose extension.
This tool allows the alignment of any two text selections, which are passed via
two --query parameters and take a query in STAMQL. Alternatively, if you want
to align two resources (a common scenario), you can just use the --resource
parameter, twice, as a more convenient shortcut.
Example invocation:
# first we create a store and add a two resource
$ stam init --resource text1.txt --resource text2.txt my.store.stam.json
# then we start the alignment (will be written to the annotation store)
$ stam align --verbose --resource text1.txt --resource text2.txt my.store.stam.json
With the --verbose flag, the alignment will be outputted to standard output in a simple TSV format with offsets for either sides, example excerpt:
/tmp/218.txt 1373-1439 /tmp/hoof001hwva02_01_0231.txt 1282-1348 "betoonen als dat van Weesp daer ick bij citatie in persoon tegens " "betoonen als dat van Weesp daer ick bij citatie in persoon tegens "
/tmp/218.txt 1444-1508 /tmp/hoof001hwva02_01_0231.txt 1348-1412 "hem begost ende wijder voor heb te procederen tot alsulke peenen" "hem begost ende wijder voor heb te procederen tot alsulke peenen"
You can also output transpositions and other alignments using the stam export --alignments (or stam query --alignments). This will output the same as
above, except for an extra first column with the annotation (transposition) ID,
and an extra final column with all annotations ID underlying the transposition
(separated by a pipe character).
stam batch
The stam batch tool is used when you want to execute multiple subcommands in
series.
Subcommands are read from standard input, either interactively or by piping input. The syntax for the subcommands is equivalent to their invocation from the command line, but with the following differences:
- there is no
stamcommand, just start with the subcommand - you can not pass input/output arguments to load/save from/to annotation stores with the individual subcommands anymore, instead, these should be passed on the batch level as a whole.
The annotation store(s) is loaded once at the start, and saved at the end if
there are any changes (and you didn't set --dry-run). This gives stam batch
its edge over just running the stam command itself in sequence; data need not
be loaded and stored after each step.
stam fromxml
The stam fromxml tool allows to map XML files with inline annotations to
STAM. It will effectively untangle the inline annotations and produce plain
text on the one hand, and stand-off STAM annotations on that plain text on the
other hand.
As there is an endless variety of XML formats imaginable, this tool takes as extra input an external configuration file that defines how to map from a specific XML format (e.g. xHTML, TEI or PageXML) to STAM. See for example this configuration for xHTML which contains some inline documentation to help you get going.
Example:
$ stam fromxml --inputfile tests/test.html --config config/fromxml/html.toml --force-new output.stam.json
Some notes:
- If you want to map HTML to STAM, first make sure your document is valid XHTML and uses the proper XML namespace. Plain HTML is not supported.
- This tool does not support conversion of stand-off annotations
formulated in XML, such as are present for instance in
FoLiA. For that format, a dedicated
stam2foliaconverter is available as part of foliatools.
stam split
With stam split you can split an annotation store by removing resources, data sets or annotations. The items to be removed are specified via the --query parameter (multiple allowed). The default behaviour is to remove the selected items, but you can reverse the behaviour by passing --keep; then all non-matching items will be removed. Use with --output to set an output filename if you don't want to overwrite and truncate your input store.
Instead of passing full queries, you can also pass resources or datasets directly via respectively --resource and --dataset.
Dependencies
~12–24MB
~363K SLoC