17 releases (10 stable)
| 2.2.4 | Nov 1, 2021 |
|---|---|
| 2.2.3 | Oct 7, 2021 |
| 2.2.2 | Jul 19, 2021 |
| 1.2.0 | Jul 17, 2021 |
| 0.1.0 | Jul 17, 2021 |
#1027 in Algorithms
28 downloads per month
30KB
391 lines
pso-rs
An easy-to-use, simple Particle Swarm Optimization (PSO) implementation in Rust.
Works in Rust 2021 edition
It uses the rand crate for random initialization, and the rayon crate for parallel objective function computation. It also has a nice progress bar curtesy of the indicatif crate. Below is a screenshot of PSO running, attempting to minimize the Lennard-Jones potential energy in a cluster of 20 molecules:
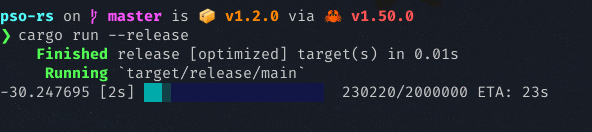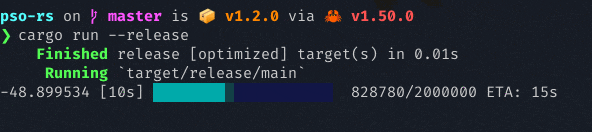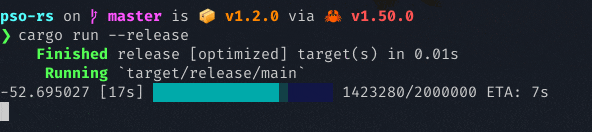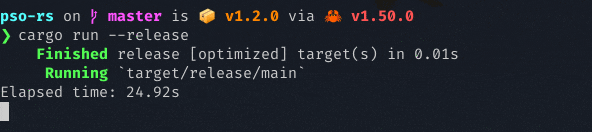
The examples below can get you started.
In order to use it in your own optimization problem, you will need to define an objective function as it is defined in the run function, and a Config object. See the Notes section for more tips.
Examples
Run PSO
use pso_rs::*;
// define objective function (d-dimensional Rosenbrock)
fn objective_function(
p: &Particle,
_flat_dim: usize,
dimensions: &Vec<usize>
) -> f64 {
(0..dimensions[0] - 1).map(|i| {
100.0 * ((p[i+1]-p[i]).powf(2.0)).powf(2.0)
+ (1.0-p[i]).powf(2.0)
}).sum()
}
// define a termination condition (optional)
fn terminate(f_best: f64) -> bool {
f_best < 1e-4
}
let config = Config {
// dimension shape of each particle
dimensions: vec![2],
// problem bounds in each dimension
bounds: vec![(-5.0, 10.0); 2],
// maximum no. of objective function computations
t_max: 10000,
// leave the rest of the params as default
..Config::default()
};
let pso = pso_rs::run(
config,
objective_function,
Some(terminate)
).unwrap();
let model = pso.model;
println!("Model: {:?} ", model.get_f_best());
Initialize PSO for later execution
use pso_rs::*;
// define objective function (d-dimensional Rosenbrock)
fn objective_function(
p: &Particle,
_flat_dim: usize,
dimensions: &Vec<usize>
) -> f64 {
(0..dimensions[0] - 1).map(|i| {
100.0 * ((p[i+1]-p[i]).powf(2.0)).powf(2.0)
+ (1.0-p[i]).powf(2.0)
}).sum()
}
let config = Config {
dimensions: vec![2],
bounds: vec![(-5.0, 10.0); 2],
t_max: 10000,
..Config::default()
};
let mut pso = pso_rs::init(
config,
objective_function
).unwrap();
// run PSO with no termination condition
pso.run(|_| false);
let model = pso.model;
println!("Model: {:?} ", model.get_f_best());
Notes
Performance
This implementation uses a flat vector (Vec<f64>) to represent any d-dimensional problem (see the Optimization Problem Dimensionality section). This means that the vector has an O(1) access time, and can be cached for fast access, similarly to a static array.
The computation of the objective function for each particle is performed in parallel, as it is computationally expensive for any non-trivial problem. In the future, complete swarms will be able to be run in parallel and optionally communicate their best found positions by passing messages.
Optimization Problem Dimensionality
Even though you can have particles of any shape and size, as long as each item is f64, pso_rs represents each particle as a flat vector: Vec<f64>.
This means that, for example, in order to find clusters of 20 molecules in 3D space that minimize the Lennard-Jones potential energy, you can define dimensions as (20, 3).
If you want, you can also create a custom reshape function, like this one for molecule clusters below:
use pso_rs::*;
fn reshape(
particle: &Particle,
particle_dims: &Vec<usize>
) -> Vec<Vec<f64>> {
let mut reshaped_cluster = vec![];
let mut i = 0;
for _ in 0..particle_dims[0] {
let mut reshaped_molecule = vec![];
for _ in 0..particle_dims[1] {
reshaped_molecule.push(particle[i]);
i += 1;
}
reshaped_cluster.push(reshaped_molecule);
}
reshaped_cluster
}
// used in the objective function
fn objective_function(
p: &Particle,
_flat_dim: usize,
dimensions: &Vec<usize>
) -> f64 {
let _reshaped_particle = reshape(p, dimensions);
/* Do stuff */
0.0
}
let config = Config {
dimensions: vec![20, 3],
bounds: vec![(-2.5, 2.5); 3],
t_max: 1,
..Config::default()
};
let pso = pso_rs::run(
config,
objective_function,
None
).unwrap();
// somewhere in main(), after running PSO as in the example:
println!(
"Best found minimizer: {:#?} ",
reshape(&pso.model.get_x_best(),
&pso.model.config.dimensions)
);
Meta
Christos A. Zonios – @czonios – c.zonios (at) uoi (dot) gr
Distributed under the MIT license. See LICENSE for more information.
https://github.com/czonios/pso-rs
Contributing
- Fork it (https://github.com/czonios/pso-rs/fork)
- Create your feature branch (
git checkout -b feature/fooBar) - Run the tests
- Commit your changes (
git commit -am 'Add some fooBar') - Push to the branch (
git push origin feature/fooBar) - Create a new Pull Request
Testing:
cargo test
cargo test --doc
When developing a new feature, it might be useful to have an optimization problem for quick feedback. You can use one of the example optimization problems (found in the src/bin directory) as follows:
# Rosenbrock function (3 dimensional)
cargo run --bin=main
# For profiling use --release flag
# You can change the number of dimensions in the file
# src/bin/main.rs
cargo run --release --bin=main
# Lennard-Jones potential for a N-particle system (N=20)
cargo run --bin=e_lj
cargo run --release --bin=e_lj
Dependencies
~4–12MB
~137K SLoC