4 releases
| 0.1.4 | Apr 8, 2020 |
|---|---|
| 0.1.3 | Nov 18, 2019 |
| 0.1.2 | Sep 23, 2019 |
| 0.1.1 | Sep 22, 2019 |
| 0.1.0 |
|
#2429 in Data structures
35 downloads per month
175KB
861 lines
nanoset.py 
A memory-optimized wrapper for Python sets likely to be empty.
⏱️ TL;DR
Save up to 85% of the memory used by empty set instances in your code with
a single line of code:
from nanoset import PicoSet as set
🚩 Table of Contents
🗺️ Overview
🐏 About Python memory usage
Python is a great programming language (fight me), but sometimes you start
questioning why it does things in certain ways. Since Python 2.3, the standard
library provides the set
collection, which is a specialized container for membership testing. On the
contrary to the ubiquitous list
collection, set is not ordered (or, more accurately, does not let you access
the order it stores the elements in). The other feature of set is that just
like its mathematical counterpart, it does not allow duplicates, which is very
useful for some algorithms. However, sets are memory-expensive:
>>> import sys
>>> sys.getsizeof(list())
72
>>> sys.getsizeof(set())
232
An empty set takes more than three times the memory of an empty list! For some
data structures or objects with a lot of set attributes, they can quickly
cause a very large part of the memory to be used for nothing. This is even more
sad when you are used to Rust, where most
collections allocate lazily.
This is where nanoset comes to the rescue:
>>> import nanoset
>>> sys.getsizeof(nanoset.NanoSet())
56
>>> sys.getsizeof(nanoset.PicoSet())
24
Actually, that's a lie, but keep reading.
💡 Simple example usecase
Let's imagine we are building an ordered graph data structure, where we may want to store taxonomic data, or any other kind of hierarchy. We can simply define the graphs and its nodes with the two following classes:
class Graph:
root: Node
nodes: Dict[str, Node]
class Node:
neighbors: Set[node]
This makes adding an edge and querying for an edge existence between two nodes
an O(1) operation, and iterating over all the nodes an O(n) operation, which
is mot likely what we want here. We use set an dnot list because we want to
avoid storing an edge in duplicate, which is a sensible choice. But now let's
look at the statistics
of the NCBITaxon project, the
database for Organismal Classification developed by the US National Center for
Biotechnology Information:
Metrics
Number of classes*: 1595237
Number of individuals: 0
Number of properties: 0
Classes without definition: 1595237
Classes without label: 0
Average number of children: 12
Classes with a single child: 40319
Maximum number of children: 41761
Classes with more than 25 children: 0
Classes with more than 1 parent: 0
Maximum depth: 38
Number of leaves**: 1130671
According to these, we are going to have 1,130,671 leaves for a total of 1,595,237 nodes, which means 70.8% of empty sets. Now you may think:
Ok, I got this. But in this case, I just need a special case for leaves, where instead of storing an empty set of
neighbors, I store a reference toNonewhen that set would be empty. I can then replace that reference with an actual set only when I want to add new edges from that node. Problem solved!
Well, glad we are on the same level: this is what nanoset does for you!
🔨 Implementation
Actually, it's not magic at all. Just imagine a class NanoSet that works as
a proxy
to an actual Python set it wraps, but which is only allocated when some data
actually needs to be stored:
class NanoSet(collections.abc.Set):
def __init__(self, iterable=None):
self.inner = None if iterable is None else set(iterable)
def add(self, element):
if self.inner is None:
self.inner = set()
self.inner.add(element)
# ... the rest of the `set` API ...
That's about it! However, doing it like so in Python would not be super
efficient, as the resulting object would be 64 bytes. Using
slots, this can be
reduced to 56 bytes, which is on par to what we get with NanoSet.
Note that these values are only when the inner set is empty! When actually
allocating the set to store our values, we allocate an additional 232 bytes
of data. This means that using NanoSet creates an overhead, since a
non-empty set will now weigh 288 bytes (256 bytes for PicoSet).
Well, I was way better off with my approach of storing
Optional[Set]everywhere then, I don't want to pay any additional cost for nonempty sets!
Sure. But that would mean changing your whole code. And actually, you may not
gain that much memory from doing that compared to using nanoset, since the
only time the wrapper performs badly is when you have a load factor of more than
90%. Furthermore, just to give you some perspective, sys.getsizeof(1) is
24 bytes as well.
By the way, you didn't mention
PicoSet. How did you manage to get that down to 24 bytes, when a slotted Python object can't be less that 56 bytes?
Easy: PicoSet is basically NanoSet, but without an implementation of the
Garbage Collector protocol.
This saves us 32 bytes of object memory, but comes with a drawback: the
garbage collector cannot see the set allocated inside the PicoSet. This
does not change anything for execution, but debugging with a memory profiler
will be harder. Here is an example where we allocate 1,000,000 singletons
first with NanoSet, then with PicoSet, using
guppy3 to check the heap:
>>> l = [nanoset.NanoSet({x}) for x in range(1000000)]
>>> guppy.hpy().heap()
Partition of a set of 3034170 objects. Total size = 328667393 bytes.
Index Count % Size % Cumulative % Kind (class / dict of class)
0 1000041 33 232100648 71 232100648 71 set
1 1000000 33 56000000 17 288100648 88 nanoset.NanoSet
...
3 96 0 8712752 3 324838712 99 list
...
>>> l = [nanoset.PicoSet({x}) for x in range(1000000)]
>>> guppy.hpy().heap()
Partition of a set of 2034285 objects. Total size = 300668995 bytes.
Index Count % Size % Cumulative % Kind (class / dict of class)
0 1000000 97 24000000 65 24000000 65 nanoset.PicoSet
1 96 0 8712752 24 32712752 89 list
...
On the second run, we have about the same order of allocated memory, saving
28 MB (28 bytes saved by switching from NanoSet to PicoSet times
1,000,000 instances). However, the garbage collector has no idea where
some of the memory is, because PicoSet hides the sets it allocates (this is
fine: it will be deallocated along with the PicoSet).
As such, I'd advise avoiding using PicoSet when debugging, which can be done
easily with Python's __debug__ flag:
if __debug__:
from nanoset import NanoSet as set
else:
from nanoset import PicoSet as set
This will cause PicoSet to be used instead of NanoSet when running Python
with the -O flag.
📈 Statistics
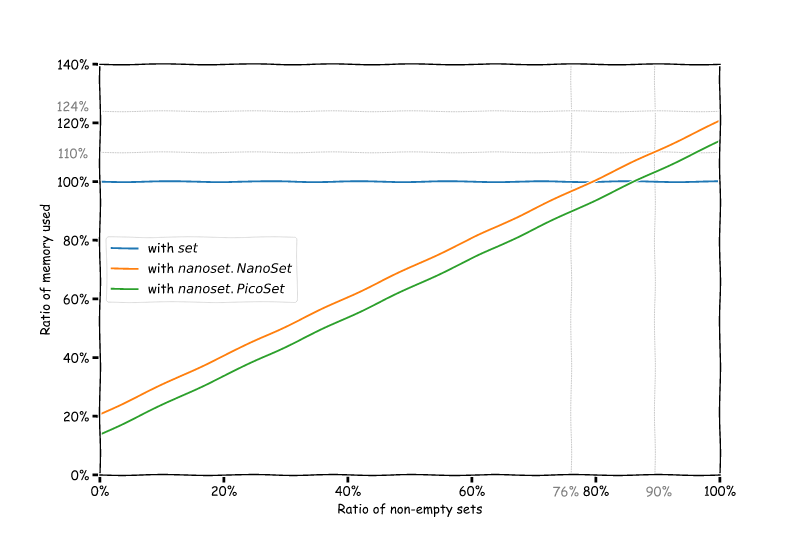
Okay, so let's do some maths. With S = 232 the size of an allocated set,
s the size of the wrapper (56 for NanoSet, 24 for PicoSet), the
x percentage of nonempty sets in our data structure, the relative size
of our sets is:
- if we're using
set: S * x / (S * x) = 100% (we use that as a reference) - if we're using
nanoset: ((S + s) * x + s * (100 - x)) / (S * x)
This gives us the following graph, which shows how much memory you can save depending of the ratio of empty sets you have at runtime:
If we get back to our NCBITaxon example, we have a total of 1,595,237 nodes
and 1,130,671 leaves, which means that by using sets we are allocating
1,595,237 * 232 = 353.0 MiB of memory simply for set after the whole
taxonomy is loaded. If we use NanoSet however, we
can reduce this to 188.0 MiB, or even to 139.3 MiB with PicoSet!
We just saved about 50% memory just by using NanoSet in place of set.
🔧 Installing
This module is implemented in Rust, but native Python wheels are compiled for the following platforms:
- Windows x86-64: CPython 3.5, 3.6, 3.7, 3.8
- Linux x86-64: CPython 3.5, 3.6, 3.7, 3.8
- OSX x86-64: CPython 3.5, 3.6, 3.7, 3.8
If you platform is not among these, you will need a
working Rust nightly toolchain
as well as the setuptools-rust
library installed to build the extension module.
Then, simply install with pip:
$ pip install --user nanoset
📖 API Reference
Well, this is a comprehensive wrapper for set, so you can just read the
standard library documentation.
Except for some very particular edge-cases, NanoSet and PicoSet both pass the
set test suite
of CPython.
There are however things you can't do:
- Subclassing a
PicoSetor aNanoSet. - Weakrefing a
PicoSetor aNanoSet. - Checking for membership in a plain
setorfrozensetwith implicit conversion tofrozenset. - Creating a
dictfrom aPicoSetor aNanoSetwithout rehashing keys.
📜 License
This library is provided under the open-source MIT license.
Dependencies
~4.5MB
~102K SLoC