5 releases (breaking)
| 0.6.0 | May 30, 2023 |
|---|---|
| 0.4.0 | Jun 2, 2022 |
| 0.3.0 | Dec 28, 2021 |
| 0.2.0 | Dec 27, 2021 |
| 0.1.0 | Dec 23, 2021 |
#1204 in Machine learning
Used in 2 crates
(via coco-rs)
1.5MB
30K
SLoC
numbbo/coco: Comparing Continuous Optimizers
Please click here to get started (unless for submitting an issue or for working on the code base)
Caution
We are currently refactoring the coco code base to make it more accessible.
Much of the documentation is therefore outdated or in a state of flux.
The code has been refactored into several repositories under github/numbbo.
We try our best to update everything as soon as possible, if you find something that you think is outdated or needs a better description, don't hestitate to open an issue or a pull request!
Important
This repository contains the source files for the coco framework.
If you don't want to extend the framework, you probably don't need this!
See instead the new documentation and use the language bindings of your choice from the package repository for your language (e.g. PyPI for Python, crates.io for Rust, ...).
[BibTeX] cite as:
Nikolaus Hansen, Dimo Brockhoff, Olaf Mersmann, Tea Tusar, Dejan Tusar, Ouassim Ait ElHara, Phillipe R. Sampaio, Asma Atamna, Konstantinos Varelas, Umut Batu, Duc Manh Nguyen, Filip Matzner, Anne Auger. COmparing Continuous Optimizers: numbbo/COCO on Github. Zenodo, DOI:10.5281/zenodo.2594848, March 2019.
This code provides a platform to
benchmark and compare continuous optimizers, AKA non-linear solvers
for numerical optimization. It is fully written in ANSI C and
Python (reimplementing the original Comparing Continous
Optimizer platform) with other languages calling the C code.
Languages currently available to connect a solver to the benchmarks are
C/C++JavaMATLABOctavePythonRust
Contributions to link further languages (including a better
example in C++) are more than welcome.
The general project structure is shown in the following figure where the black color indicates code or data provided by the platform and the red color indicates either user code or data and graphical output from using the platform:
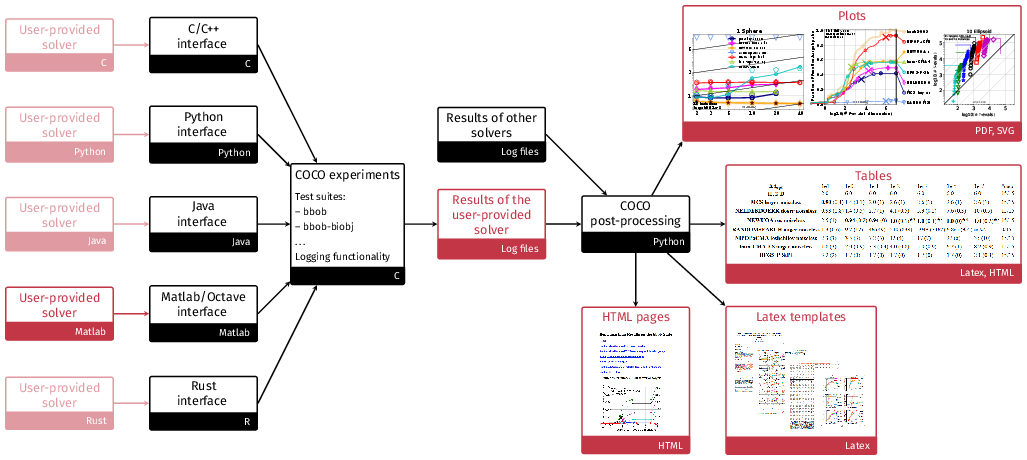
For more general information:
- The GitHub.io documentation pages for COCO
- The article on benchmarking guidelines and an introduction to COCO: A Platform for Comparing Continuous Optimizers in a Black-Box Setting (pdf) or at arXiv
- The COCO experimental setup description
- The BBOB workshops series
- For COCO/BBOB news register here
- See links below to learn even more about the ideas behind COCO
Getting Started
Running Experiments
-
For running experiments in Python, follow the (new) instructions here.
Otherwise, download the COCO framework code from github,- either by clicking the Download ZIP button
and unzip the
zipfile, - or by typing
git clone https://github.com/numbbo/coco.git. This way allows to remain up-to-date easily (but needsgitto be installed). After cloning,git pullkeeps the code up-to-date with the latest release.
The record of official releases can be found here. The latest release corresponds to the master branch as linked above.
- either by clicking the Download ZIP button
and unzip the
-
In a system shell,
cdinto thecocoorcoco-<version>folder (framework root), where the filedo.pycan be found. Type, i.e. execute, one of the following commands oncepython do.py run-c python do.py run-java python do.py run-matlab python do.py run-octave python do.py run-pythondepending on which language shall be used to run the experiments.
run-*will build the respective code and run the example experiment once. The build result and the example experiment code can be found undercode-experiments/build/<language>(<language>=matlabfor Octave).python do.pylists all available commands. -
Copy the folder
code-experiments/build/YOUR-FAVORITE-LANGUAGEand its content to another location. In Python it is sufficient to copy the fileexample_experiment_for_beginners.pyorexample_experiment2.py. Run the example experiment (it already is compiled). As the details vary, see the respective read-me's and/or example experiment files:Cread me and example experimentJavaread me and example experimentMatlab/Octaveread me and example experimentPythonread me and example experiment2
If the example experiment runs, connect your favorite algorithm to Coco: replace the call to the random search optimizer in the example experiment file by a call to your algorithm (see above). Update the output
result_folder, thealgorithm_nameandalgorithm_infoof the observer options in the example experiment file.Another entry point for your own experiments can be the
code-experiments/examplesfolder. -
Now you can run your favorite algorithm on the
bbobandbbob-largescalesuites (for single-objective algorithms), on thebbob-biobjsuite (for multi-objective algorithms), or on the mixed-integer suites (bbob-mixintandbbob-biobj-mixintrespectively). Output is automatically generated in the specified dataresult_folder. By now, more suites might be available, see below.
Post-processing Data
-
Install the post-processing for displaying data (using Python):
pip install cocopp -
Postprocess the data from the results folder of a locally run experiment by typing
python -m cocopp [-o OUTPUT_FOLDERNAME] YOURDATAFOLDER [MORE_DATAFOLDERS]Any subfolder in the folder arguments will be searched for logged data. That is, experiments from different batches can be in different folders collected under a single "root"
YOURDATAFOLDERfolder. We can also compare more than one algorithm by specifying several data result folders generated by different algorithms. -
We also provide many archived algorithm data sets. For example
python -m cocopp 'bbob/2009/BFGS_ros' 'bbob/2010/IPOP-ACTCMA'processes the referenced archived BFGS data set and an IPOP-CMA data set. The given substring must have a unique match in the archive or must end with
!or*or must be a regular expression containing a*and not ending with!or*. Otherwise, all matches are listed but none is processed with this call. For more information in how to obtain and display specific archived data, seehelp(cocopp)orhelp(cocopp.archives)or the classCOCODataArchive.Data descriptions can be found for the
bbobtest suite at coco-algorithms and for thebbob-biobjtest suite at coco-algorithms-biobj. For other test suites, please see the COCO data archive.Local and archived data can be freely mixed like
python -m cocopp YOURDATAFOLDER 'bbob/2010/IPOP-ACT'which processes the data from
YOURDATAFOLDERand the archived IPOP-ACT data set in comparison.The output folder,
ppdataby default, contains all output from the post-processing. Theindex.htmlfile is the main entry point to explore the result with a browser. Data from the same foldername as previously processed may be overwritten. If this is not desired, a different output folder name can be chosen with the-o OUTPUT_FOLDERNAMEoption.A summary pdf can be produced via LaTeX. The corresponding templates can be found in the
code-postprocessing/latex-templatesfolder. Basic html output is also available in the result folder of the postprocessing (filetemplateBBOBarticle.html). -
In order to exploit more features of the post-processing module, it is advisable to use the module within a Python or IPython shell or a Jupyter notebook or JupyterLab, where
import cocopp help(cocopp)provides the documentation entry pointer.
If you detect bugs or other issues, please let us know by opening an issue in our issue tracker at https://github.com/numbbo/coco/issues.
Known Issues / Trouble-Shooting
Post-Processing
Too long paths for postprocessing
It can happen that the postprocessing fails due to too long paths to the algorithm data. Unfortunately, the error you get in this case does not indicate directly to the problem but only tells that a certain file could not be read. Please try to shorten the folder names in such a case.
Font issues in PDFs
We have occasionally observed some font issues in the pdfs, produced by the postprocessing
of COCO (see also issue #1335). Changing to
another matplotlib version solved the issue at least temporarily.
BibTeX under Mac
Under the Mac operating system, bibtex seems to be messed up a bit with respect to
absolute and relative paths which causes problems with the test of the postprocessing
via python do.py test-postprocessing. Note that there is typically nothing to fix if
you compile the LaTeX templates "by hand" or via your LaTeX IDE. But to make the
python do.py test-postprocessing work, you will have to add a line with
openout_any = a to your texmf.cnf file in the local TeX path. Type
kpsewhich texmf.cnf to find out where this file actually is.
Algorithm appears twice in the figures
Earlier versions of cocopp have written extracted data to a folder named _extracted_....
If the post-processing is invoked with a * argument, these folders become an argument and
are displayed (most likely additionally to the original algorithm data folder). Solution:
remove the _extracted_... folders and use the latest version of the post-processing
module cocopp (since release 2.1.1).
Implementation Details
-
The C code features an object oriented implementation, where the
coco_problem_tis the most central data structure / object.coco.h,example_experiment.candcoco_internal.hare probably the best pointers to start to investigate the code (but see also here).coco_problem_tdefines a benchmark function instance (in a given dimension), and is called viacoco_evaluate_function. -
Building, running, and testing of the code is done by merging/amalgamation of all C-code into a single C file,
coco.c, andcoco.h. (by callingdo.py, see above). Like this it becomes very simple to include/use the code in different projects. -
Cython is used to compile the C to Python interface in
build/python/interface.pyx. The Python module installation filesetup.pyuses the compiledinterface.c, ifinterface.pyxhas not changed. For this reason, Cython is not a requirement for the end-user.
Citation
You may cite this work in a scientific context as
N. Hansen, A. Auger, R. Ros, O. Mersmann, T. Tušar, D. Brockhoff. COCO: A Platform for Comparing Continuous Optimizers in a Black-Box Setting, Optimization Methods and Software, 36(1), pp. 114-144, 2021. [pdf, arXiv]
@ARTICLE{hansen2021coco,
author = {Hansen, N. and Auger, A. and Ros, R. and Mersmann, O.
and Tu{\v s}ar, T. and Brockhoff, D.},
title = {{COCO}: A Platform for Comparing Continuous Optimizers
in a Black-Box Setting},
journal = {Optimization Methods and Software},
doi = {https://doi.org/10.1080/10556788.2020.1808977},
pages = {114--144},
issue = {1},
volume = {36},
year = 2021
}
Links About the Workshops and Data
- The BBOB workshop series, which uses the NumBBO/Coco framework extensively, can be tracked here
- Data sets from previous experiments for many algorithms are available at
- https://numbbo.github.io/data-archive/bbob/ for the
bbobtest suite - https://numbbo.github.io/data-archive/bbob-noisy/ for the
bbob-noisytest suite - https://numbbo.github.io/data-archive/bbob-biobj/ for the
bbob-biobjtest suite, and at - https://numbbo.github.io/data-archive/bbob-largescale/ for the
bbob-largescaletest suite.
- https://numbbo.github.io/data-archive/bbob/ for the
- Postprocessed data for each year in which a BBOB workshop was taking place can be found at https://numbbo.github.io/ppdata-archive
- Stay informed about the BBOB workshop series and releases of the NumBBO/Coco software by registering via this form
- Downloading this repository
- via the above green "Clone or Download" button or
- by typing
git clone https://github.com/numbbo/coco.gitor - via https://github.com/numbbo/coco/archive/master.zip in your browser
Comprehensive List of Documentations
-
General introduction: COCO: A Platform for Comparing Continuous Optimizers in a Black-Box Setting (pdf) or at arXiv
-
Experimental setup: http://numbbo.github.io/coco-doc/experimental-setup/
-
Testbeds
bbob: https://numbbo.github.io/gforge/downloads/download16.00/bbobdocfunctions.pdfbbob-biobj: http://numbbo.github.io/coco-doc/bbob-biobj/functions/bbob-biobj-ext: http://numbbo.github.io/coco-doc/bbob-biobj/functions/bbob-noisy(only in old code basis): https://hal.inria.fr/inria-00369466/document/bbob-largescale: https://arxiv.org/pdf/1903.06396.pdfbbob-mixint: https://hal.inria.fr/hal-02067932/documentbbob-biobj-mixint: https://numbbo.github.io/gforge/preliminary-bbob-mixint-documentation/bbob-mixint-doc.pdfbbob-constrained(in progress): http://numbbo.github.io/coco-doc/bbob-constrained
-
Performance assessment: http://numbbo.github.io/coco-doc/perf-assessment/
-
Performance assessment for biobjective testbeds: http://numbbo.github.io/coco-doc/bbob-biobj/perf-assessment/
-
APIs
Cexperiments code: http://numbbo.github.io/coco-doc/C- Python experiments code (module
cocoex): https://numbbo.github.io/coco-doc/apidocs/cocoex - Python short experiment code example for beginners
- Python
example_experiment2.py: https://numbbo.github.io/coco-doc/apidocs/example - Postprocessing code (module
cocopp): https://numbbo.github.io/coco-doc/apidocs/cocopp
-
Somewhat outdated documents:
- Former home page: https://web.archive.org/web/20210504150230/https://coco.gforge.inria.fr/
- Full description of the platform: http://coco.lri.fr/COCOdoc/
- Experimental setup before 2016: http://coco.lri.fr/downloads/download15.03/bbobdocexperiment.pdf
- Old framework software documentation: http://coco.lri.fr/downloads/download15.03/bbobdocsoftware.pdf
-
Some examples of results.
Dependencies
~0–2.2MB
~44K SLoC