3 unstable releases
Uses old Rust 2015
| 0.2.1 | Feb 9, 2021 |
|---|---|
| 0.2.0 | Feb 9, 2021 |
| 0.1.0 | Dec 31, 2016 |
#1690 in Database interfaces
1MB
806 lines
UniParc XML parser
- Introduction
- Usage
- Table schema
- Installation
- Output files
- Benchmarks
- Roadmap
- FAQ (Frequently Asked Questions)
- FUQ (Frequently Used Queries)
Introduction
Process the UniParc XML file (uniparc_all.xml.gz) downloaded from the UniProt website into CSV files that can be loaded into a relational database.
Usage
Uncompressed XML data can be piped into uniparc_xml_parser in order to
$ curl -sS ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/uniparc/uniparc_all.xml.gz \
| zcat \
| uniparc_xml_parser
The output is a set of CSV (or more specifically TSV) files:
$ ls
-rw-r--r-- 1 user group 174G Feb 9 13:52 xref.tsv
-rw-r--r-- 1 user group 149G Feb 9 13:52 domain.tsv
-rw-r--r-- 1 user group 138G Feb 9 13:52 uniparc.tsv
-rw-r--r-- 1 user group 107G Feb 9 13:52 protein_name.tsv
-rw-r--r-- 1 user group 99G Feb 9 13:52 ncbi_taxonomy_id.tsv
-rw-r--r-- 1 user group 74G Feb 9 20:13 uniparc.parquet
-rw-r--r-- 1 user group 64G Feb 9 13:52 gene_name.tsv
-rw-r--r-- 1 user group 39G Feb 9 13:52 component.tsv
-rw-r--r-- 1 user group 32G Feb 9 13:52 proteome_id.tsv
-rw-r--r-- 1 user group 15G Feb 9 13:52 ncbi_gi.tsv
-rw-r--r-- 1 user group 21M Feb 9 13:52 pdb_chain.tsv
-rw-r--r-- 1 user group 12M Feb 9 13:52 uniprot_kb_accession.tsv
-rw-r--r-- 1 user group 656K Feb 9 04:04 uniprot_kb_accession.parquet
Table schema
The generated CSV files conform to the following schema:
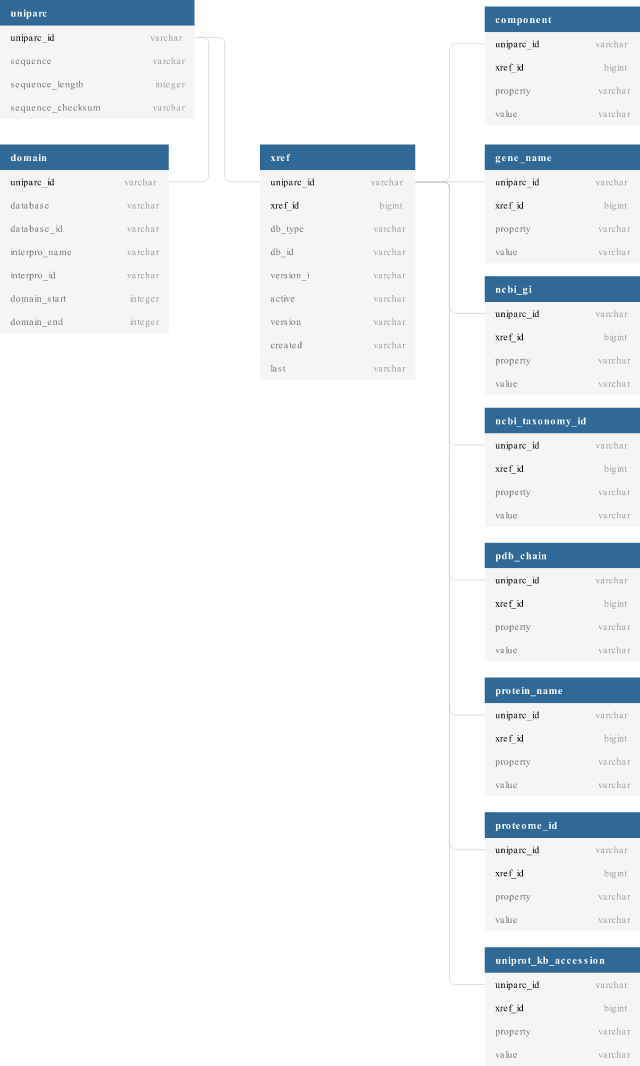
Installation
Binaries
Linux binaries are available here: https://gitlab.com/ostrokach/uniparc_xml_parser/-/packages.
Cargo
Use cargo to compile and install uniparc_xml_parser for your target platform:
cargo install uniparc_xml_parser
Conda
Use conda to install precompiled binaries:
conda install -c ostrokach-forge uniparc_xml_parser
Output files
Parquet
Parquet files containing the processed data are available at the following URL and are updated monthly: http://uniparc.data.proteinsolver.org/.
Google BigQuery
The data can also be queried directly using Google BigQuery: https://console.cloud.google.com/bigquery?project=ostrokach-data&p=ostrokach-data&page=dataset&d=uniparc.
Benchmarks
Parsing 10,000 XML entires takes around 30 seconds (the process is mostly IO-bound):
$ time bash -c "zcat uniparc_top_10k.xml.gz | uniparc_xml_parser >/dev/null"
real 0m33.925s
user 0m36.800s
sys 0m1.892s
The actual uniparc_all.xml.gz file has around 373,914,570 elements.
Roadmap
- Keep everything in bytes all the way until output.
FAQ (Frequently Asked Questions)
Why not split uniparc_all.xml.gz into multiple small files and process them in parallel?
- Splitting the file requires reading the entire file. If we're reading the entire file anyway, why not parse it as we read it?
- Having a single process which parses
uniparc_all.xml.gzmakes it easier to create an incremental unique index column (e.g.xref.xref_id).
FUQ (Frequently Used Queries)
TODO
Dependencies
~9–15MB
~260K SLoC