2 unstable releases
| 0.2.0 | Jun 24, 2022 |
|---|---|
| 0.1.1 | Feb 12, 2022 |
#622 in Machine learning
1MB
1K
SLoC
affinityprop
The affinityprop crate provides an optimized implementation of the Affinity Propagation
clustering algorithm, which identifies cluster of data without a priori knowledge about the
number of clusters in the data. The original algorithm was developed by
Brendan Frey and Delbery Dueck
About
Affinity Propagation identifies a subset of representative examples from a dataset, known as exemplars.
Briefly, the algorithm accepts as input a matrix describing pairwise similarity for all data
values. This information is used to calculate pairwise responsibility and availability.
Responsibility r(i,j) describes how well-suited point j is to act as an exemplar for
point i when compared to other potential exemplars. Availability a(i,j) describes how
appropriate it is for point i to accept point j as its exemplar when compared to
other exemplars.
Users provide a number of convergence iterations to repeat the calculations, after which the potential exemplars are extracted from the dataset. Then, the algorithm continues to repeat until the exemplar values stop changing, or until the maximum iterations are met.
Why this crate?
The nature of Affinity Propagation demands an O(n2) runtime. An existing sklearn version is implemented using the Python library numpy which incorporates vectorized row operations. Coupled with SIMD instructions, this results in decreased time to finish.
However, in applications with large input values, the O(n2) runtime is still prohibitive. This crate implements Affinity Propagation using the rayon crate, which allows for a drastic decrease in overall runtime - as much as 30-60% when compiled in release mode!
Dependencies
cargo
with rustc >=1.58
Installation
In Rust code
[dependencies]
affinityprop = "0.2.0"
ndarray = "0.15.4"
As a command-line tool
cargo install affinityprop
Usage
From Rust code
The affinityprop crate expects a type that defines how to calculate pairwise Similarity
for all data points. This crate provides the NegEuclidean, NegCosine, and
LogEuclidean structs, which are defined as -1 * sum((a - b)**2), -1 * (a . b)/(|a|*|b|),
and sum(log((a - b)**2)), respectively.
Users who wish to calculate similarity differently are advised that Affinity Propagation expects s(i,j) > s(i, k) iff i is more similar to j than it is to k.
use ndarray::{arr1, arr2, Array2};
use affinityprop::{AffinityPropagation, NegCosine, Preference};
let x: Array2<f32> = arr2(&[[0., 1., 0.], [2., 3., 2.], [3., 2., 3.]]);
// Cluster using negative cosine similarity with a pre-defined preference
let ap = AffinityPropagation::default();
let (converged, results) = ap.predict(&x, NegCosine::default(), Preference::Value(-10.));
assert!(converged && results.len() == 1 && results.contains_key(&0));
// Cluster with list of preference values
let pref = arr1(&[0., -1., 0.]);
let (converged, results) = ap.predict(&x, NegCosine::default(), Preference::List(&pref));
assert!(converged);
assert!(results.len() == 2 && results.contains_key(&0) && results.contains_key(&2));
// Use damping=0.5, threads=2, convergence_iter=10, max_iterations=100,
// median similarity as preference
let ap = AffinityPropagation::new(0.5, 2, 10, 100);
let (converged, results) = ap.predict(&x, NegCosine::default(), Preference::Median);
assert!(converged);
assert!(results.len() == 2 && results.contains_key(&0) && results.contains_key(&2));
// Predict with pre-calculated similarity
let s: Array2<f32> = arr2(&[[0., -3., -12.], [-3., 0., -3.], [-12., -3., 0.]]);
let ap = AffinityPropagation::default();
let (converged, results) = ap.predict_precalculated(s, Preference::Value(-10.));
assert!(converged && results.len() == 1 && results.contains_key(&1));
From the Command Line
affinityprop can be run from the command-line and used to analyze a file of data:
ID1 val1 val2
ID2 val3 val4
ID3 val5 val6
where IDn is any string identifier and valn are floating-point (decimal) values.
The file delimiter is provided from the command line with the -l flag.
Similarity will be calculated based on the option set by the -s flag.
For files without row ids:
val1 val2
val3 val4
val5 val6
provide the -n flag from the command line. IDs will automatically be assigned by zero-based
index.
Users may instead provide a pre-calculated similarity matrix by passing the -s 3 flag from
the command line and by structuring their input file as:
ID1 sim11 sim12 sim13
ID2 sim21 sim22 sim23
ID3 sim31 sim32 sim33
where rowi, colj is the pairwise similarity between inputs i and j.
Or, for files without row labels, users may pass -n -s 3:
sim11 sim12 sim13
sim21 sim22 sim23
sim31 sim32 sim33
IDs will automatically be assigned by zero-based index.
Help Menu
affinityprop 0.2.0
Chris N. <christopher.neely1200@gmail.com>
Vectorized and Parallelized Affinity Propagation
USAGE:
affinityprop [OPTIONS] --input <INPUT>
FLAGS:
-n, --no_labels Input file does not contain IDS as the first column
-h, --help Prints help information
-V, --version Prints version information
OPTIONS:
-c, --convergence_iter <CONV_ITER> Convergence iterations, default=10
-d, --damping <DAMPING> Damping value in range (0, 1), default=0.9
-l, --delimiter <DELIMITER> File delimiter, default '\t'
-i, --input <INPUT> Path to input file
-m, --max_iter <MAX_ITER> Maximum iterations, default=100
-r, --precision <PRECISION> Set f32 or f64 precision, default=f32
-p, --preference <PREF> Preference to be own exemplar, default=median pairwise similarity
-s, --similarity <SIMILARITY> Set similarity calculation method
(0=NegEuclidean,1=NegCosine,2=LogEuclidean,3=precalculated), default=0
-t, --threads <THREADS> Number of worker threads, default=4
Results
Results are printed to stdout in the format:
Converged=true/false nClusters=NC nSamples=NS
>Cluster=n size=N exemplar=i
[comma-separated cluster member IDs/indices]
>Cluster=n size=N exemplar=i
[comma-separated cluster member IDs/indices]
...
Runtime and Resource Notes
Affinity Propagation is O(n2) in both runtime and memory. This crate seeks to address the former, not the latter.
An estimated memory usage can be calculated given:
memory(GB) = p * 4 * N^2 / 2^30
For N inputs. p = 4 for 32-bit floating-point precision and p = 8 for 64-bit.
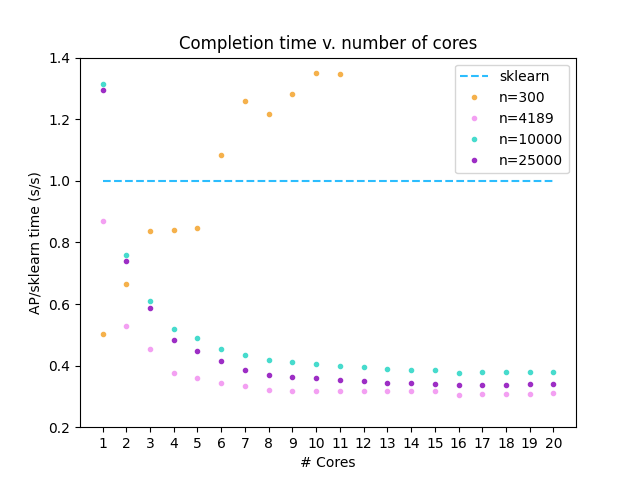
Comparison to sklearn implementation
This implementation was tested on 50-D isotropic Gaussian blobs generated by the sklearn
make_blobs
function (n=300,10000,25000). A dataset of biological relevance
was also selected (n=4189). ARI/F1 scores >= 0.99 were obtained for the Gaussian data when compared to the labels
provided by make_blobs. ARI/F1 = 0.98 was obtained for the biological dataset when compared to sklearn-derived
labels.
This affinityprop implementation was compared against the Affinity Propagation
implementation contained within scikit-learn-1.0.2, and run using numpy-1.22.2 with Python 3.10.0.
This analysis was completed using a Ryzen 9 3950X processor.
In all analyses, damping=0.95, convergence_iter=400, and max_iter=4000. Preference=-1000.0 for Gaussian data and -10.0 for biological data.
Dependencies
~4.5MB
~78K SLoC