3 releases
| 0.1.2 | Apr 30, 2024 |
|---|---|
| 0.1.1 | Apr 30, 2024 |
| 0.1.0 | Apr 30, 2024 |
#83 in Testing
341 downloads per month
62KB
1K
SLoC
Pymute: A Mutation Testing Tool for Python/Pytest written in Rust
Pymute is inspired by my experience of using cargo mutants. I used it in a rust project, really enjoyed it, so I had to search for something similar for python/pytest projects. Quickly found mut.py and the pynguin fork of mut.py among some other solutions, but none of them seemed quite stable across different python versions.
Pymute takes a somewhat naive approach and simply creates a temporary directory
for every mutant, and then runs pytest in that working directory independent
of other mutations (note that if pymute is interrupted, some manual clean up
of /tmp may be required). Mutations are inserted by simply manipulating the text in the
*.py files rather than operating on the AST and therefore should work across most versions.
Installation
To install pymute make sure you have rust and cargo installed on your system
(check cargo --version). You can follow instructions here to install rust and
its toolchain: https://www.rust-lang.org/tools/install
You can install pymute via cargo crom crates.io.
cargo install pymute
Alternatively, you can install pymute via cargo from GitHub as:
cargo install --git https://github.com/LeSasse/pymute.git
Verify the correctness of the installation using pymute --version or pymute --help.
How to run it:
Pymute allows you to run your tests on mutants using two different runners:
If you are using pytest (which is also the default runner), then pymute assumes that your pytest tests can be run from the root directory of your python project and that they can be run in independent copies of your python project without failing. For example if you have a project in:
~/projects/my_project
and you copy this as:
cp -r ~/projects/my_project /tmp/my_project
Then you should be able to run the tests in the copy as:
cd /tmp/my_project
python -m pytest .
Importantly, since this approach does not take care of setting up environments or
installing your package, it is necessary that the python -m pytest invocation
tests against the local copy and not the installed version, so that
the tests run correctly for each mutant. This should generally
be the case if you are using a src-less layout (see https://blog.ionelmc.ro/2014/05/25/python-packaging/
for the definition of src-less layout) according to pytest docs,
so that running from the root of the project using the python -m pytest . invocation
will actually run the test against the local module and NOT the installed version.
However, pytest and imports can be quite confusing, and if you are not sure, you
can actually run the tests using tox. This will create a virtual environment and install
your package for each mutant separately, so you can be sure the tests run correctly
against each mutant version of your package. Importantly, you do not need
to run all the tox environments but using the --environment option you can run specific
tox environments. Overall, of course, this approach will be considerably slower though
due to having to set up all the tox environments.
Example
This repository comes with a small example of a python project with some basic tests. You can test this out by cloning this repo if you like:
# set up an environment with pytest and tox if you dont have it already
python -m venv .env
source .env/bin/activate
pip install tox pytest
git clone https://github.com/LeSasse/pymute.git
cd pymute
pymute example
This should give you the following output:
[MISSED] Mutant Survived: + replaced by - in file example/src/model.py on line 6
[MISSED] Mutant Survived: 5 replaced by 6 in file example/src/model.py on line 16
[MISSED] Mutant Survived: 0 replaced by 1 in file example/src/model.py on line 17
[MISSED] Mutant Survived: 0 replaced by 1 in file example/src/model.py on line 22
[MISSED] Mutant Survived: == replaced by != in file example/src/model.py on line 27
By default, pymute only shows mutants that were missed, i.e. mutants for which
your tests all passed. This is most informative because it tells you that these
or similar bugs could have been introduced to your program without your tests alerting you.
These replacements change the behaviour of the program, i.e. represent a bug/regression,
but the tests passed anyways. In other words, the tests were unable to safeguard
the project from these bugs and they could have been checked into the main branch.
There are two more output levels though, caught and process. You can specify them as:
pymute example --output-level caught
pymute example --output-level process
The caught level will also print out mutants that your tests caught successfully,
so that the bug could not have been introduced. The process level will also print
out all the output from the underlying pytest or tox processes. This is useful
for verifying that the processes are actually running correctly (for example, maybe you
forgot to activate the correct environment and pytest or tox is not actually installed).
This is important since pymute will only check if a process was successful or not.
On a Bigger Project
Let's put pymute to the test using a larger python project: julearn.
Running long test suites for potentially hundreds of mutants may not be feasible with the approach that pymute takes. Therefore pymute provides the options to mutate only subsets of your program and to run only subsets of your tests.
Let's first try and run pymute on the whole project. We can use pytest since
julearn uses a src-less layout and so therefore the python -m pytest . invocation
should run tests on local modules rather than the installed one.
# set up an environment with pytest and tox if you dont have it already
python -m venv .env
source .env/bin/activate
pip install tox pytest
git clone https://github.com/juaml/julearn.git
cd julearn
# we can install it to install all the dependencies
pip install ".[docs,deslib,viz,skopt,dev]"
We can set the number of threads to control the number of mutants running in parallel.
Keep in mind that each thread will need some disk space in your /tmp so you should
consider this, so that threads don't fail because you are running out of space in /tmp.
We can set the output level to caught to get a bit more output about whats happening
and run it as:
pymute . --output-level caught --num-threads 4
However, this finds more than a thousand mutants and seems to mutate files in docs
and other folders that are not actually part of the package. pymute will look
for mutants anywhere under the root of your project (i.e. pymute's first positional
argument). Instead, we can be a bit more specific by providing the --modules
option. This is a glob expression that will specify that pymute should only look
for mutants in files that match it. Importantly, pymute will automatically filter
out files that start with "test_" and files that end with *_test.py to avoid
creating mutations for pytest tests. It is also important to wrap the glob
expression in a string, so that its not actually interpreted as a glob expression
by your shell but handed over to pymute as a string.
pymute . --output-level caught --num-threads 4 --modules "julearn/**/*.py"
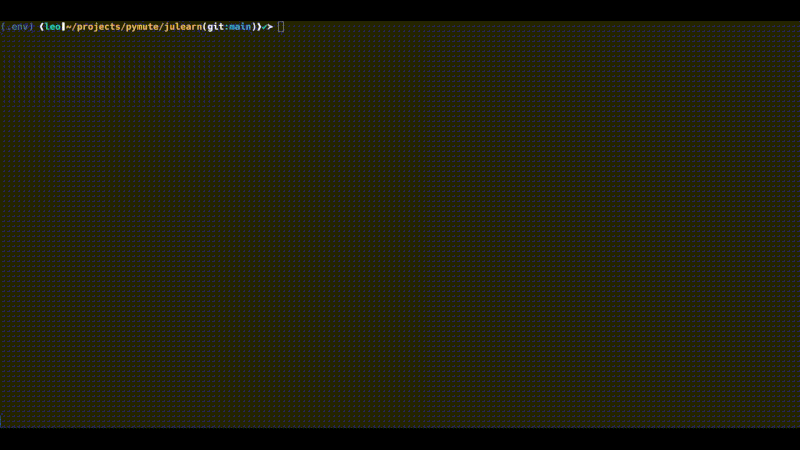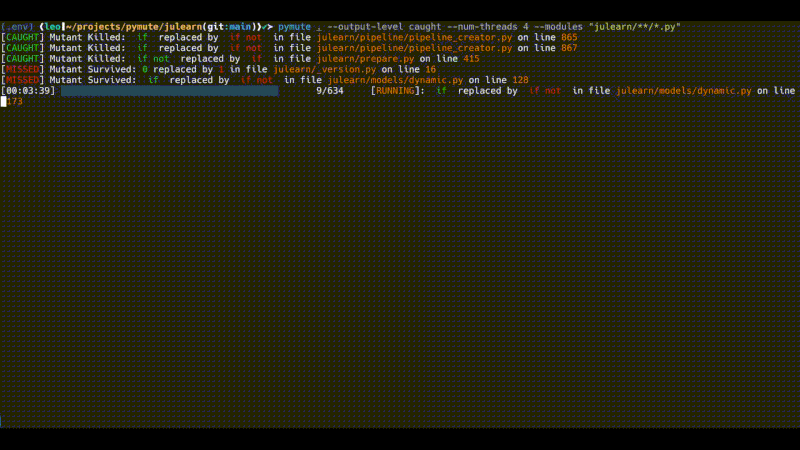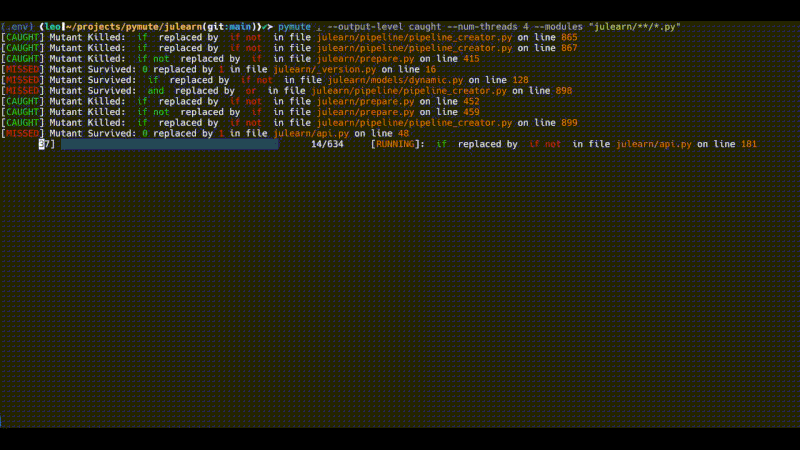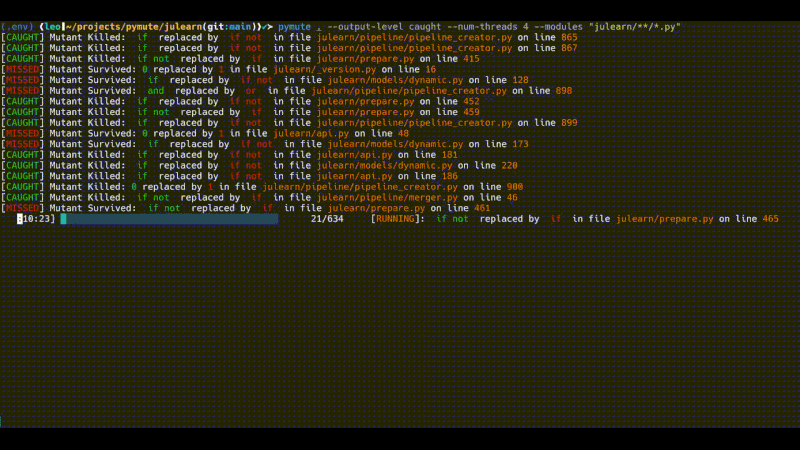
However, this still finds some 600 mutants and runs quite slowly. The output above was running for about 10 minutes (the gif is sped up). There are a number of ways to further subset the mutants, or to subset the tests that are run in order to perform more specific testing, that doesn't take as much time.
Run a Random Subset of Mutants across the whole Package
You can run a randomly sampled subset of mutants across the whole package by specifying
the --max-mutants option. Each individual test run will still be slow, but there
are less to do overall, so that pymute will finish sooner:
pymute . --output-level caught --num-threads 4 --modules "julearn/**/*.py" --max-mutants 10

This command took a bit less than 5 minutes (gif is sped up), and while it found some
interesting MISSED mutations, each run still takes quite a bit of time.
Running Specific Tests for Mutants in Specific Modules (RECOMMENDED WAY of using pymute)
Often, you just want to focus on improving tests for a specific module, and
so running the whole test suite is a waste of time. You create or change some module
and you then want to perform mutation testing for the tests that specifically are meant
to catch regressions in these modules. You can do this by specififying the --tests
option, which will run python -m pytest for only these tests. For example, we
might focus on the modules in julearn/model_selection. We can run this as:
pymute . \
--output-level caught \
--num-threads 4 \
--modules "julearn/model_selection/*.py" \
--tests julearn/model_selection/tests

This run finished in 20 seconds and the gif finally did not have to be sped up
to show some interesting output. We can now easily and quickly inspect the MISSED
mutants and investigate how they would have changed the behaviour of some public API
of that module and whether we can better test for such changed behaviour. This approach
is recommended when using pymute because it allows for much quicker iteration
of mutation runs. You can improve the tests and then run pymute again with the
same command and it should go quite fast.
Subset the Mutation Types
One further way to subset the mutants that pymute will run is by specifying the
--mutation-types option. This is a list of types separated by commas. The help
text (pymute --help) gives the following options:
--mutation-types <MUTATION_TYPES>
Mutation types
[default: math-ops conjunctions booleans control-flow comp-ops numbers]
Possible values:
- math-ops: Mutate mathematical operators (e.g. "*,+,-,/")
- conjunctions: Mutate conjunctions in boolean expressions (e.g. "and/or")
- booleans: Mutate booleans (e.g. "True/False")
- control-flow: Mutate control flow statements (e.g. if statements)
- comp-ops: Mutate comparison operators (e.g. "<,>,==,!=")
- numbers: Mutate numbers (e.g. off-by-one errors)
So for example to only mutate numbers and comparison operators, we could run the previous
command with the following --mutation-types option (gif is also NOT sped up):
pymute . \
--output-level caught \
--num-threads 4 \
--modules "julearn/model_selection/*.py" \
--tests julearn/model_selection/tests \
--mutation-types numbers,comp-ops

Dependencies
~9–20MB
~291K SLoC